Biophysics
Vol.
10
No.
03
(
2022
), Article ID:
54780
,
7
pages
10.12677/BIPHY.2022.103005
SHISAL1基因真核表达质粒构建 及其在肝癌细胞中表达
吴文昊,周莉莉,石静,严辉,陈帅,孙达权*
贵州医科大学基础医学院,贵州 贵阳
收稿日期:2022年7月27日;录用日期:2022年8月3日;发布日期:2022年8月15日

摘要
本文的目的是克隆人SHISAL1基因和构建其真核表达载体,并在肝细胞癌细胞中表达和分析。以人肝细胞THLE-3总RNA为模板,用RT-PCR获得SHISAL1基因蛋白编码区,通过基因克隆和重组获得真核表达载体pDsRed1-SHISAL1;用脂质体介导法将pDsRed1-SHISAL1导入肝癌细胞HuH-7中,用荧光显微镜和激光共聚焦扫描显微镜观察细胞内红色荧光,用蛋白免疫印迹和免疫荧光染色分析细胞内FZD3的表达情况。结果显示成功克隆人SHISAL1基因蛋白编码区并获得了真核表达质粒pDsRed1-SHISAL1;荧光显微术和蛋白免疫印迹结果显示外源SHISAL1融合蛋白,即SHISAL1-RFP蛋白,在肝癌细胞HuH-7中表达;激光共聚焦扫描显微术和蛋白免疫印迹结果显示SHISAL1可以抑制肝癌细胞HuH-7内源性FZD3表达。本研究成功克隆了SHISAL1基因并在肝癌细胞中表达,并证明SHISAL1影响肝癌细胞内源性FZD3表达。
关键词
SHISAL1基因,基因克隆,真核表达,肝癌细胞

Construction of Eukaryotic Expressional Plasmid of SHISAL1 Gene and Its Expression in Hepatocellular Carcinoma Cells
Wenhao Wu, Lili Zhou, Jing Shi, Hui Yan, Shuai Chen, Daquan Sun*
College of Basic Medical Sciences, Guizhou Medical University, Guiyang Guizhou
Received: Jul. 27th, 2022; accepted: Aug. 3rd, 2022; published: Aug. 15th, 2022

ABSTRACT
The aims are to clone human SHISAL1 gene and construct its eukaryotic expressional vector, and then to express and analyze the protein in hepatocellular carcinoma cells. Using THLE-3 total RNA as a template, the protein coding region of SHISAL1 gene was obtained by RT-PCR, and the eukaryotic expressional vector pDsRed1-SHISAL1 was obtained by gene cloning and recombination. The recombinant plasmid was introduced into hepatocellular carcinoma cell line HuH-7 by liposome method. The intracellular red fluorescence was observed by fluorescence microscopy and laser confocal scanning microscopy, and the expression of endogenous FZD3 was analyzed by western blot and immunofluorescence staining. The results showed that the protein coding region of human SHISAL1 gene was cloned successfully and the eukaryotic expression plasmid pDsRed1-SHISAL1 was obtained. Fluorescence microscopy and western blot showed that SHISAL1 fusion protein, namely SHISAL1-RFP fusion protein, was expressed in HuH-7 cells. Laser confocal scanning microscopy and western blot showed that overexpression of SHISAL1 could inhibit the endogenous expression of FZD3 in HuH-7 cells. Collectively, SHISAL1 gene was successfully cloned and expressed in hepatocellular carcinoma cells, and then it was proved that SHISAL1 could affect the expression of endogenous FZD3 in hepatocellular carcinoma cells.
Keywords:SHISAL1 Gene, Gene Cloning, Eukaryotic Expression, Hepatocellular Carcinoma Cell

Copyright © 2022 by author(s) and Hans Publishers Inc.
This work is licensed under the Creative Commons Attribution International License (CC BY 4.0).
http://creativecommons.org/licenses/by/4.0/


1. 引言
人SHISAL1基因全名为Shisa like 1,也叫KIAA1644,已确定定位于人22号常染色体短臂的q13.31上。与许多传统发现的基因相比,它的发现是通过生物信息学数据库搜索和比对获得的,现已证实SHISAL1基因编码的蛋白质且属于SHISA超家族成员之一,推测其可能是一种跨膜适配器并可能具备多种生物学功能 [1] [2]。生物信息学分析发现SHISAL1的N-末端是一个长约25个氨基酸的信号肽序列,其后是一段富含6个半胱氨酸的保守结构域(C * C * CC * CC),在整个蛋白结构的中间是一段跨膜结构域,其后带有含正电荷的精氨酸/赖氨酸残基,C-末端含有一段富含脯氨酸的结构域 [1]。本组前期生物信息学分析发现SHISAL1可能与多种肿瘤进程密切相关。因此,在此之前本组已通过大肠杆菌原核表达系统获取了SHISAL1全蛋白,并通过小鼠免疫制备了SHISAL1多克隆抗体,准备研究SHISAL1蛋白表达、定位及生物学功能研究 [2]。为揭示SHISAL1在肝癌发生、发展和浸润中的具体生物学功能及作用途径,本组在此先克隆重组了SHISAL1基因的真核表达质粒,通过脂质体介导法导入并获得了表达外源SHISAL1的肝细胞癌细胞,并分析发现外源SHISAL1可以抑制肝细胞癌细胞内源性FZD3的表达,为揭示SHISAL1在肝癌进程中的作用奠定一定的前期基础。
2. 实验材料和方法
2.1. 实验材料
肝细胞癌细胞HuH-7来源于中科院上海细胞库,SV40转化的肝上皮细胞系THLE-3来源于上海冠导生物工程有限公司,两株细胞均经过STR鉴定。脂质体LipofectamineTM 2000、细胞培养液、TRIzol试剂购买于赛默飞世尔科技(中国)有限公司。胎牛血清来源于Gemini公司。限制性核酸内切酶、cDNA第一链合成试剂盒、高保真Taq DNA聚合酶、3'末端加“A”试剂盒、DNA胶纯化试剂盒、T4 DNA连接酶来源于宝日医生物技术(北京)有限公司。引物合成和DNA测序由上海生工生物科技有限公司完成。RFP标签鼠单抗、FZD3多克隆兔抗、GAPDH多克隆兔抗来源于Bioworld。SHISAL1多克隆鼠抗由本实验室自行制备。
2.2. 细胞培养
SV40转化的肝上皮细胞系THLE-3细胞和肝细胞癌细胞系HuH-7细胞用分别用含10%胎牛血清的RPMI-1640培养液和DMEM培养液进行培养,细胞培养的条件为37℃、5.0% CO2。当细胞生长融合至80%~90%时用0.25%胰酶液进行消化传代和收集。
2.3. RNA抽提和RT-PCR
将细胞THLE-3播种于直径6 cm的细胞培养皿上,待细胞生长融合至80%时,弃去培养液,用PBS清洗细胞3次,然后在培养皿中直接加入1 mL的TRIzol试剂,反复吹打裂解细胞,按照TRIzol说明书的操作程序提取细胞总RNA。测定RNA溶液浓度,用反转录试剂盒在42℃反转录合成cDNAs。
2.4. SHISAL1基因引物设计和克隆
根据GeneBank中SHISAL1基因序列号(NM_001099294)设计人SHISAL1基因蛋白编码区两端的PCR引物,并在两端末端引入下游操作所需的特异性限制性核酸内切酶的酶切位点,引物序列如表1所示。
Table 1. PCR primers for human SHISAL1 gene coding region
表1. 人SHISAL1基因蛋白编码区的PCR引物
2.5. 真核表达载体构建
以人SHISAL1基因PCR为引物,以THLE-3细胞的cDNAs为模板,用高保真PCR法扩增SHISAL1基因的蛋白编码区,其中设置退火温度为62℃,延伸时间为30 s。PCR产物用1.5%的琼脂糖凝胶电泳鉴定和纯化。纯化的DNA片段通过加末端加“A”试剂盒对DNA片段3’末端加“A”,加“A”产物与T-载体pMD18-T连接,形成重组质粒pMD18T-SHISAL1。连接产物用热激法转化感受态细菌DH5α,通过含氨苄青霉素的LB 固体培养基筛选培养转化的细菌。挑取单克隆菌落并在LB细菌培养液中扩大培养,抽提质粒和双酶切鉴定,鉴定符合预期效果的质粒进行DNA测序分析。
测序正确的重组质粒pMD18T-SHISAL1和真核表达载体pDsRed1-N1用EcoR I/Xho I进行双酶切,酶切产物用1.5%琼脂糖凝胶电泳分离纯化SHISAL1 DNA片段和线性化骨架载体,胶回收试剂盒回收并纯化目的片段并用T4 DNA连接酶连接,产物转化感受态细菌DH5α,用含有卡那霉素的LB 固体培养基筛选培养转化的细菌,挑取单克隆菌落并扩大培养,经抽提质粒、双酶切鉴定和DNA 测序,正确重组的真核表达质粒命名为pDsRed1-SHISAL1。
2.6. 细胞转染
将HuH-7细胞在转染前12 h按30%~40%的融合率铺板于6孔板中。转染前将细胞培养液更换为无血清的DMEM,转染过程和转染试剂用量按LipofectamineTM 2000说明书进行。实验设置对照组(空载体pDsRed1-N1)和实验组(目的基因质粒pDsRed1-SHISAL1)。转染后6 h更换为新鲜的含10% FBS的细胞培养液继续培养,转染后72 h将细胞放置在倒置荧光显微镜下观察转染效果。
2.7. 蛋白免疫印迹
用0.25%的胰酶消化培养的靶细胞,离心收集后用PBS清洗2次。用高效细胞裂解液快速裂解细胞并进行BCA定量,5 ´ SDS上样缓冲液制备样品后进行变性胶电泳,将蛋白通过电转移印至PVDF膜上,5% BSA室温封闭PVDF膜30 min,TBST清洗3次,4℃一抗孵育过夜,TBST清洗3次,二抗室温孵育2 h,TBST清洗3次,ECL显色和成像。
2.8. 免疫荧光染色
用0.25%的胰酶消化细胞,重新铺板于底层垫有盖玻片(多聚赖氨酸包被)的24孔板中,当靶细胞贴壁并显示正常的细胞形态后,弃去细胞培养液,PBS清洗盖玻片3次,用4%的多聚甲醛4℃固定过夜,用0.5% Triton X-100室温通透细胞10 min,5% BSA室温封闭30 min,一抗4℃孵育过夜、二抗室温孵育2 h、DAPI染色和封片,激光扫描共聚焦显微镜分析细胞内的蛋白质分布与表达。
3. 结果
3.1. 成功克隆人SHISAL1基因和建立其真核表达载体pDsRed1-SHISAL1
如图1所示,通过RT-PCR和高保真PCR扩增获得人SHISAL1基因的蛋白编码区cDNA片段(图1A),将该片段插入克隆载体pMD18-T中,经EcoR I/Xho I双酶切鉴定(图1B)和DNA测序,结果显示SHISAL1蛋白编码区无任何氨基酸突变。将此编码区DNA片段插入到真核表达载体pDsRed1-N1中,构建的重组质粒pDsRed1-SHISAL1先后进行双酶切鉴定(图1C)和DNA测序,结果显示成功构建重组真核表达质粒,在细胞中可以表达SHISAL1融合蛋白(SHISAL1-RFP),其重组质粒结构示意图如图1D所示。
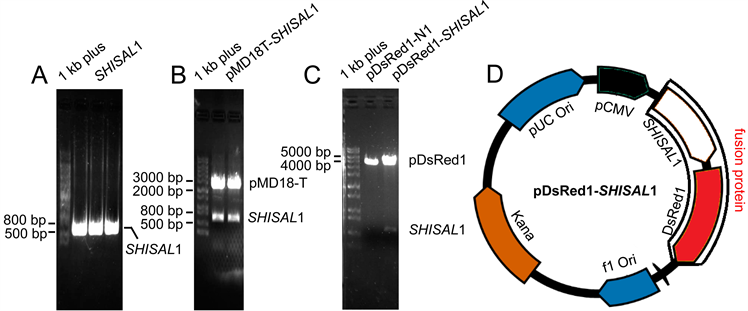 注:(A) SHISAL1基因蛋白编码区cDNA。(B) EcoR I/Xho I酶切分析pMD18T-SHISAL1。(C) EcoR I/Xho I酶切分析重组质粒pDsRed1-SHISAL1。(D) 真核表达载体pDsRed1-SHISAL1结构示意图。
注:(A) SHISAL1基因蛋白编码区cDNA。(B) EcoR I/Xho I酶切分析pMD18T-SHISAL1。(C) EcoR I/Xho I酶切分析重组质粒pDsRed1-SHISAL1。(D) 真核表达载体pDsRed1-SHISAL1结构示意图。
Figure 1. Gene coding region of SHISAL1 and its eukaryotic expressional plasmid
图1. SHISAL1基因蛋白编码区及其真核表达载体
3.2. 外源SHISAL1在肝癌细胞HuH-7中表达
如图2A所示,用脂质体将重组质粒pDsRed1-SHISAL1导入肝癌细胞HuH-7中72小时后,细胞在荧光显微镜下开始出现红色荧光。对细胞进行爬片、固定和激光共聚焦扫描分析,发现细胞内红色荧光呈局部和区域性分布(图2B)。对细胞进行蛋白免疫印迹分析,发现用SHISAL1抗体孵育后,对照细胞仅出现内源性SHISAL1一条条带,而转染组细胞出现了2条条带;用RFP标签抗体进行孵育,对照细胞出现了红色荧光蛋白(RFP)一条条带,而转染组细胞出现了一条分子量较大的RFP融合蛋白(SHISAL1-RFP) (图C),上述结果证明外源SHISAL1蛋白在肝癌细胞HuH-7中表达。
 注:(A) 外源SHISAL1蛋白在荧光显微镜下呈红色。(B) 外源SHISAL1蛋白在激光共聚焦扫描显微镜下呈红色荧光。(C) Western blot显示外源SHISAL1蛋白在HuH-7中表达。
注:(A) 外源SHISAL1蛋白在荧光显微镜下呈红色。(B) 外源SHISAL1蛋白在激光共聚焦扫描显微镜下呈红色荧光。(C) Western blot显示外源SHISAL1蛋白在HuH-7中表达。
Figure 2. Expression of SHISAL1 protein in HuH-7 cells of hepatocellular carcinoma
图2. 外源蛋白SHISAL1在肝癌细胞HuH-7中表达
3.3. 表达外源SHISAL1蛋白抑制肝癌细胞HuH-7中FZD3蛋白的表达
当肝癌细胞HuH-7中表达外源SHISAL1后,对细胞用FZD3多克隆抗体进行细胞免疫荧光染色分析,结果发现肝癌细胞表达外源SHISAL1后,细胞内FZD3表达下降(图3A)。用蛋白免疫印迹对细胞内全蛋白进行分析,同样发现过表达外源SHISAL1后,细胞内源性FZD3表达下降(图3B),证明SHISAL1可能会抑制肝癌细胞HuH-7中内源性FZD3的表达。
 注:(A) 激光共聚焦扫描显微术显示SHISAL1抑制细胞内FZD3表达。(B) 蛋白免疫印迹显示SHISAL1抑制FZD3的表达。
注:(A) 激光共聚焦扫描显微术显示SHISAL1抑制细胞内FZD3表达。(B) 蛋白免疫印迹显示SHISAL1抑制FZD3的表达。
Figure 3. Suppression of expression of FZD3 in HuH-7 cells by SHISAL1
图3. SHISAL1抑制细胞HuH-7中FZD3的表达
4. 讨论
目前已发现的Shisa超家族包括Shisa家族、Shisa like家族两大类,前者成员包括SHISA 1~9,后者包括Shisa的远缘同源基因Shisa like 1 (SHISAL1)、Shisa like 2a (SHISAL2a)和Shisa like 2b (SHISAL2b),其共同特征是含有一段富含半胱氨酸的保守的序列 [1] [3]。Shisa超家族成员广泛存在于脊椎动物中,与胚胎发育 [4]、成体心肌细胞重编程 [5]、肌肉细胞融合 [6]、糖尿病 [7]、细胞凋亡 [8]、神经发育与传导 [9] [10]、神经疾病 [11] [12]、细胞干性维持 [13] 和肿瘤病变 [14] [15] 等均有密切联系。但是,目前尚缺乏对Shisa like 家族的探索和研究,作为Shisa like家族中的SHISAL1,科研工作者对其了解也仅停留在转录和翻译水平上。最新研究发现在年轻的乳腺癌患者或正常乳腺组织中,若包括SHISAL1基因在内的5个基因集表达过高就会导致总体生存率和无病生存率显著下降 [16];可是在子宫内膜癌中呈现相反的趋势,即当子宫内膜癌患者组织中高表达包括SHISAL1基因在内的7个差异表达基因集,其预后效果显著提高 [17]。本组通过Ualcan网站对TCGA数据库中的35种肿瘤进行分析,发现有数据的26种原发性肿瘤中,10种肿瘤中SHISAL1表达中位值升高(其中7种差异显著),16种肿瘤中SHISAL1表达中位值下降(其中9种差异显著) [18]。这些结果暗示SHISAL1生物学功能的复杂性,它可能在不同的组织癌变中起着不同的调控作用,也显示当前我们对SHISAL1缺乏系统和全面的认识。但是在蛋白水平上,目前仅了解SHISAL1基因可以翻译成蛋白质并定位于高尔基体上,其他具体的生物学功能知之甚少 [2]。
为研究SHISAL1在肝癌发生、发展、转移和浸润中的相关作用以及其所扮演的角色,本课题组以肝细胞癌细胞HuH-7为研究对象,通过体外克隆和构建SHISAL1基因的红色荧光蛋白融合的真核表达载体,将其导入肝癌细胞中,使其在肝癌细胞中可以定位、追踪和分析蛋白生物学功能,发现其过表达后可以抑制肝癌细胞内源性FZD3表达,目前已知FZD3是一个典型的与胚胎发育和肿瘤发生发展密切相关的原癌基因,其表达升高会导致多种肿瘤发生、发展和转移 [19] [20] [21]。因此,本组推测SHISAL1在肝癌进程中可能会通过抑制FZD3表达而抑制细胞癌变,但其中涉及的作用机制和途径有待进一步研究和探索。
致谢
本项目所使用的大型仪器设备均由贵州医科大学基础医学科学研究中心提供。
基金项目
本研究由国家自然科学基金(项目编号:81560390)和2020年贵州省大学生创新创业训练计划项目(项目编号:S202010660004和342)提供资助。
文章引用
吴文昊,周莉莉,石 静,严 辉,陈 帅,孙达权. SHISAL1基因真核表达质粒构建及其在肝癌细胞中表达
Construction of Eukaryotic Expressional Plasmid of SHISAL1 Gene and Its Expression in Hepatocellular Car-cinoma Cells[J]. 生物物理学, 2022, 10(03): 39-45. https://doi.org/10.12677/BIPHY.2022.103005
参考文献
- 1. Pei, J. and Grishin, N.V. (2012) Unexpected Diversity in Shisa-Like Proteins Suggests the Importance of Their Roles as Transmembrane Adaptors. Cell Signal, 24, 758-769. https://doi.org/10.1016/j.cellsig.2011.11.011
- 2. 周莉莉, 陈相屹, 余畅, 湛钊, 雷霆雯, 孙达权. 小鼠样人SHisa样蛋白1 (SSL1)多克隆抗体制备及其亚细胞定位[J]. 细胞与分子免疫学杂志, 2019, 35(9): 843-848.
- 3. Abdollahi Nejat, M., Klaassen, R.V., Spijker, S. and Smit, A.B. (2021) Auxiliary Subunits of the AMPA Receptor: The Shisa Family of Proteins. Current Opinion in Pharmacology, 58, 52-61. https://doi.org/10.1016/j.coph.2021.03.001
- 4. Onishi, K. and Zou, Y. (2017) Sonic Hedgehog Switches on Wnt/Planar Cell Polarity Signaling in Commissural Axon Growth Cones by Reducing Levels of Shisa2. Elife, 6, e25269. https://doi.org/10.7554/eLife.25269
- 5. Noack, C., Iyer, L.M., Liaw, N.Y., Schoger, E., Khadjeh, S., Wagner, E., Woelfer, M., Zafiriou, M.P., Milting, H., Sossalla, S., Streckfuss-Boemeke, K., Hasenfuß, G., Zimmermann, W.H. and Zelarayán, L.C. (2019) KLF15-Wnt-Dependent Cardiac Reprogramming Up-Regulates SHISA3 in the Mammalian Heart. JACC: Journal of the American College of Cardiology, 74, 1804-1819. https://doi.org/10.1016/j.jacc.2019.07.076
- 6. Liu, Z., Wang, C., Liu, X. and Kuang, S. (2018) Shisa2 Regulates the Fusion of Muscle Progenitors. Stem Cell Research, 31, 31-41. https://doi.org/10.1016/j.scr.2018.07.004
- 7. Kumar, P., Traurig, M. and Baier, L.J. (2020) Identification and Functional Validation of Genetic Variants in Potential miRNA Target Sites of Established BMI Genes. International Journal of Obesity (London), 44, 1191-1195. https://doi.org/10.1038/s41366-019-0488-8
- 8. Kim, N., Kim, M.J., Sung, P.S., Bae, Y.C., Shin, E.C. and Yoo, J.Y. (2016) Interferon-Inducible Protein SCOTIN Interferes with HCV Replication through the Autolysosomal Degrada-tion of NS5A. Nature Communications, 7, Article No. 10631. https://doi.org/10.1038/ncomms10631
- 9. Schmitz, L.J.M., Klaassen, R.V., Ruiperez-Alonso, M., Zamri, A.E., Stroeder, J., Rao-Ruiz, P., Lodder, J.C., van der Loo, R.J., Mansvelder, H.D., Smit, A.B. and Spijker, S. (2017) The AMPA Receptor-Associated Protein Shisa7 Regulates Hippo-campal Synaptic Function and Contextual Memory. Elife, 6, e24192. https://doi.org/10.7554/eLife.24192.030
- 10. Klaassen, R.V., Stroeder, J., Coussen, F., Hafner, A.S., Petersen, J.D., Renancio, C., Schmitz, L.J., Normand, E., Lodder, J.C., Rotaru, D.C., Rao-Ruiz, P., Spijker, S., Mansvelder, H.D., Choquet, D. and Smit, A.B. (2016) Shisa6 Traps AMPA Receptors at Postsynaptic Sites and Prevents Their Desensitiza-tion during Synaptic Activity. Nature Communications, 7, Article No. 10682. https://doi.org/10.1038/ncomms10682
- 11. Sabaie, H., Talebi, M., Gharesouarn, J., Asadi, M.R., Jalaiei, A., Arsang-Jang, S., Hussen, B.M., Taheri, M., Jalili Khoshnoud, R. and Rezazadeh, M. (2022) Identification and Analysis of BCAS4/hsa-miR-185-5p/SHISA7 Competing Endogenous RNA Axis in Late-Onset Alzheimer’s Disease Using Bi-oinformatic and Experimental Approaches. Frontiers in Aging Neuroscience, 14, Article ID: 812169. https://doi.org/10.3389/fnagi.2022.812169
- 12. Sabaie, H., Gharesouran, J., Asadi, M.R., Farhang, S., Ahangar, N.K., Brand, S., Arsang-Jang, S., Dastar, S., Taheri, M. and Rezazadeh, M. (2022) Downregulation of miR-185 Is a Common Pathogenic Event in 22q11.2 Deletion Syndrome-Related And Idiopathic Schizophrenia. Metabolic Brain Dis-ease, 37, 1175-1184. https://doi.org/10.1007/s11011-022-00918-5
- 13. Tokue, M., Ikami, K., Mizuno, S., Takagi, C., Miyagi, A., Taka-da, R., Noda, C., Kitadate, Y., Hara, K., Mizuguchi, H., Sato, T., Taketo, M.M., Sugiyama, F., Ogawa, T., Kobayashi, S., Ueno, N., Takahashi, S., Takada, S. and Yoshida, S. (2017) SHISA6 Confers Resistance to Differentiation-Promoting Wnt/β-Catenin Signaling in Mouse Spermatogenic Stem Cells. Stem Cell Reports, 8, 561-575. https://doi.org/10.1016/j.stemcr.2017.01.006
- 14. Maffei, R., Fiorcari, S., Martinelli, S., Benatti, S., Bulgarelli, J., Rizzotto, L., Debbia, G., Santachiara, R., Rigolin, G.M., Forconi, F., Rossi, D., Laurenti, L., Palumbo, G.A., Vallisa, D., Cuneo, A., Gaidano, G., Luppi, M. and Marasca, R. (2018) Increased SHISA3 Expression Characterizes Chronic Lym-phocytic Leukemia Patients Sensitive to Lenalidomide. Leukemia & Lymphoma, 59, 423-433. https://doi.org/10.1080/10428194.2017.1339872
- 15. Tamura, K., Furihata, M., Satake, H., Hashida, H., Kawada, C., Osakabe, H., Fukuhara, H., Kumagai, N., Iiyama, T., Shuin, T. and Inoue, K. (2017) SHISA2 Enhances the Aggres-sive Phenotype in Prostate Cancer through the Regulation of WNT5A Expression. Oncology Letters, 14, 6650-6658. https://doi.org/10.3892/ol.2017.7099
- 16. Paul, A.M., George, B., Saini, S., Pillai, M.R., Toi, M., Costa, L. and Kumar, R. (2022) Delineation of Pathogenomic Insights of Breast Cancer in Young Women. Cells, 11, 1927. https://doi.org/10.3390/cells11121927
- 17. Liu, L., Lin, J. and He, H. (2019) Identification of Potential Crucial Genes Associated with the Pathogenesis and Prognosis of Endometrial Cancer. Frontiers in Genetics, 10, Article No. 373. https://doi.org/10.3389/fgene.2019.00373
- 18. Chandrashekar, D.S., Bashel, B., Balasubramanya, S.A.H., Creighton, C.J., Rodriguez, I.P., Chakravarthi, B.V.S.K. and Varambally, S. (2017) UALCAN: A Portal for Facilitating Tumor Subgroup Gene Expression and Survival Analyses. Neoplasia, 19, 649-658. https://doi.org/10.1016/j.neo.2017.05.002
- 19. Zhu, G.Q., Wang, Y., Wang, B., Liu, W.R., Dong, S.S., Chen, E.B., Cai, J.L., Wan, J.L., Du, J.X., Song, L.N., Chen, S.P., Yu, L., Zhou, Z.J., Wang, Z., Zhou, J., Shi, Y.H., Fan, J. and Dai, Z. (2022) Targeting HNRNPM Inhibits Cancer Stemness and Enhances Antitumor Immunity in Wnt-Activated Hepato-cellular Carcinoma. Cellular and Molecular Gastroenterology and Hepatology, 13, 1413-1447. https://doi.org/10.1016/j.jcmgh.2022.02.006
- 20. Li, C., Nguyen, V., Clark, K.N., Zahed, T., Sharkas, S., Filipp, F.V. and Boiko, A.D. (2019) Down-Regulation of FZD3 Receptor Suppresses Growth and Metastasis of Human Mela-noma Independently of Canonical WNT Signaling. Proceedings of the National Academy of Sciences of the United States of America, 116, 4548-4557. https://doi.org/10.1073/pnas.1813802116
- 21. Wang, A., Wang, Y., Zhan, J. and Chen, J. (2021) Mi-croRNA-186-5p Inhibits the Metastasis of Cervical Cancer by Targeting FZD3. Journal of BUON, 26, 677-683.
