Advances in Clinical Medicine
Vol.
13
No.
01
(
2023
), Article ID:
60736
,
11
pages
10.12677/ACM.2023.131100
缺氧和免疫抑制的胰腺癌肿瘤 微环境中PLAU的表达及 对预后的影响
刘任政1,吴仰东1,孙 琳2,胡 骁1*
1青岛大学附属医院肝胆胰外科,山东 青岛
2青岛大学附属医院重症医学科,山东 青岛
收稿日期:2022年12月28日;录用日期:2023年1月21日;发布日期:2023年1月30日

摘要
目的:基于生物信息分析,探究PLAU (蛋白编码基因)在以缺氧和免疫抑制为特征的胰腺恶性肿瘤微环境中的表达及其对疾病的进展和预后的影响。方法:通过生物信息分析,使用R语言相关的在线数据库,研究免疫浸润和缺氧在胰腺癌发生发展中的作用,通过Gene ontology (GO)/Kyoto Encyclopedia of Genes and Genomes (KEGG)富集分析筛选参与调控胰腺癌缺氧和免疫浸润微环境的关键基因;结合UCSC XENA数据库的转录组数据(RNAseq)及随访数据分析对预后的影响;通过ssGSEA算法分析PLAU在胰腺癌免疫细胞中的表达程度和p值;同时,qRT-PCR验证该基因在正常胰腺导管细胞、Panc-1及缺氧环境下培养的Panc-1表达的差异。结果:通过生信分析及GO/KEEG途径,发现PLAU在正常组织和肿瘤组织中的表达有显著差异;根据OS曲线,发现PLAU高表达患者的生存时间明显缩短,单因素COX回归分析发现PLAU、N1期和病理期II期是胰腺癌的独立预后因素,多因素cox回归分析显示,PLAU和N1期是胰腺癌的独立预后因素;PLAU与树突状细胞、巨噬细胞、中性粒细胞、NK CD56dim细胞等免疫细胞浸润程度呈正相关,与Th17细胞浸润程度呈负相关,PLAU高表达组与低表达组相比免疫细胞分布存在差异性;PLAU在胰腺癌细胞中呈高表达,在缺氧的胰腺癌细胞中PLAU的表达低于正常培养的胰腺癌细胞。结论:PLAU在胰腺癌细胞中的差异表达提示其是一种关键的预后基因。它与胰腺癌的缺氧和免疫侵袭有关,在胰腺癌的肿瘤微环境中起重要作用。
关键词
PLAU,胰腺癌,组织缺氧,免疫浸润,预后

Expression of PLAU in Tumor Microenvironment of Hypoxic and Immunosuppressive Pancreatic Cancer and Its Influence on Prognosis
Rengzheng Liu1, Yangdong Wu1, Lin Sun2, Xiao Hu1*
1Department of Hepatobiliary Pancreatic Surgery, The Affiliated Hospital of Qingdao University, Qingdao Shandong
2Department of ICU, The Affiliated Hospital of Qingdao University, Qingdao Shandong
Received: Dec. 28th, 2022; accepted: Jan. 21st, 2023; published: Jan. 30th, 2023

ABSTRACT
Objective: Based on bioinformatics analysis, the expression of PLAU (protein coding gene) in the microenvironment of pancreatic malignant tumors characterized by hypoxia and immunosuppression and its impact on disease progression and prognosis were investigated. Methods: The role of immune invasion and hypoxia in the occurrence and development of pancreatic cancer was studied through bioinformatics analysis and R language related online database, and key genes involved in regulating the hypoxia and immune invasion microenvironment of pancreatic cancer were screened through Gene Ontology (GO)/Kyoto Encyclopedia Of Genes and Genomes (KEGG) enrichment analysis; Combined with the UCSC XENA RNAseq database and follow-up data, the impact on prognosis was analyzed; The expression and p value of PLAU in pancreatic cancer immune cells were analyzed by ssGSEA algorithm; At the same time, qRT-PCR in vitro was used to verify the difference in the expression of this gene in normal pancreatic duct cells, Panc-1 and Panc-1 cultured in hypoxia environment. Results: Through the bioinformatics analysis and GO/KEEG pathway, it was found that the expression of PLAU in normal and tumor tissues was significantly different; According to the OS curve, the survival time of patients with high PLAU expression was significantly shortened. Single factor COX regression analysis found that PLAU, N1 and pathological stage II were independent prognostic factors of pancreatic cancer, and multivariate cox regression analysis showed that PLAU and N1 were independent prognostic factors of pancreatic cancer; PLAU was positively correlated with the infiltration of dendritic cells, macrophages, neutrophils, NK CD56dim cells and other immune cells, and negatively correlated with the infiltration of Th17 cells. The distribution of immune cells in the high expression group of PLAU was different from that in the low expression group; PLAU was highly expressed in pancreatic cancer cells, and the expression of PLAU in hypoxia pancreatic cancer cells was lower than that in normal pancreatic cancer cells. Conclusion: The differential expression of PLAU in pancreatic cancer cells suggests that PLAU is a key prognostic gene. It is related to hypoxia and immune invasion of pancreatic cancer and plays an important role in the tumor microenvironment of pancreatic cancer.
Keywords:PLAU, Pancreatic Cancer, Hypoxia, Immune Infiltration, Prognosis

Copyright © 2023 by author(s) and Hans Publishers Inc.
This work is licensed under the Creative Commons Attribution International License (CC BY 4.0).
http://creativecommons.org/licenses/by/4.0/


1. 引言
胰腺癌(Pancreatic Adenocarcinoma, PAAD)是世界上预后最差的癌症之一,是全球癌症相关死亡的第四大因素 [1]。胰腺在腹腔内的特殊位置导致胰腺癌发病早期隐藏特征 [2],并且胰腺癌缺乏高度敏感的分子靶向标志物,导致了胰腺癌早期诊断的困难性 [3] [4]。胰腺癌具有很强的侵袭性,多数患者一旦确诊即是肿瘤晚期,很难找到有效的治疗方案 [5],手术是目前唯一有可能根治的治疗方法,但因为很少有胰腺癌患者可以得到早期诊断,导致手术切除几率低,且术后复发率极高,短期生存率也不容乐观。胰腺癌是对放疗敏感性较低,对无法行手术治疗或为防止术后复发的患者,目前可通过化学治疗,以期能降低手术后的复发与转移几率 [6]。目前,随着分子生物学研究在临床上应用的发展,基因治疗和免疫治疗成为了目前综合治疗中重要的一部分 [7]。虽然近年来随着对胰腺癌研究的深入,一些关于胰腺癌的诊断、放疗技术和系统治疗的研究成果不断被提出,但胰腺癌的生存率并没有明显提高,相关死亡人数仍在增加 [8]。因此,发现并提出一种潜在的高灵敏度分子靶向标记物,对胰腺癌的诊断和治疗具有重要意义。
在胰腺癌中,除去肿瘤细胞,其余主要由成纤维细胞、细胞外基质、内皮细胞和干细胞组成,这些成分构成了机体的肿瘤微环境 [7]。肿瘤的生物学功能很大程度上取决于癌细胞与微环境之间的相互作用 [9] [10]。肿瘤微环境的细胞类型有助于高度免疫抑制、缺氧和促纤维增生的癌细胞生长 [9] [11]。由于广泛的结缔组织增生继发血管减少导致组织氧含量降低是胰腺肿瘤微环境的重要特征之一 [12],缺氧环境是导致胰腺癌进展的因素之一 [13],并在多种细胞和生物事件中发挥关键作用,包括细胞增殖、代谢,新生血管生成和肿瘤生长、侵袭、转移 [14]。胰腺癌的免疫环境一般被认为是免疫抑制 [15] [16],免疫抑制因子与胰腺癌患者预后不良相关 [17] [18],肿瘤高度的免疫微环境使得对免疫治疗非常不敏感。PLAU是胰腺恶性肿瘤中的纤溶酶激活因子,在体内参与许多生理和病理过程 [19]。根据以往的研究表明,PLAU促进各种癌症的发生和进展,包括结直肠癌和食管癌 [20] [21]。PLAU在肿瘤的迁移、侵袭和转移中也起重要作用 [22] [23]。
因此,胰腺肿瘤缺氧微环境和免疫抑制对肿瘤本身的发展和预后起着重要的作用,本研究旨在通过生物信息学的方法研究胰腺癌中的特殊目标分子PLAU,探索其在胰腺癌中的表达及缺氧环境下其对胰腺肿瘤微环境中的免疫抑制的作用的影响,它提供了一个新的想法对胰腺癌的诊断和治疗。
2. 方法
2.1. 细胞培养和试剂
人胰腺癌细胞panc-1和人正常胰管细胞hTERT-HPNE购于(购自中国科学院细胞库所),于添加10%FBS和1%青霉素/链霉素的DMEM高糖培养基(均自美伦生物采购)中,在温度为37℃、CO2体积分数为5%、湿度为95%的培养箱中培养。缺氧处理的细胞在缺氧培养箱中培养24 h。
2.2. 缺氧关键基因及其表达
使用在线数据库GEPIA2 [24] 来寻找胰腺癌的差异表达基因,以logFC和P值作为差异表达基因的筛选条件。以满足P < 0.05和|log2FC| > 1作为基因表达差异的2个条件筛选目的基因,同时log2FC > 2为高表达基因,log2FC < −2为低表达基因。然后通过火山图进行差异基因可视化处理。
通过GO/KEGG富集分析筛选缺氧相关的功能基因及相关的通路。通过R.Main package cluster Profiler [3.14.3];org.Hs.eg.db package [3.10.0];GOplot package [1.0.2] 对基因组进行分析。
通过UCSC XENA数据库RNAseq (https://xenabrowser.net/datapages/)中提取胰腺癌基因组图谱,在GTEx数据库提取相应的正常组织基因图谱。通过ggplot2 [3.3.3版本],得到PLAU在胰腺癌和正常胰腺中的表达。我们还通过ggplot2分析了PLAU在泛癌中的表达,得到了PLAU在泛癌中的表达。
通过来源于human protein map (Hpa, https://www.proteinatlas.org)得到PLAU在胰腺癌组织和正常组织的免疫组化染色图像。
2.3. 生存与预后分析
通过R语言利用survival package [3.2-10]分析胰腺癌基因组,利用RNAseq中位基因表达值来区分高表达和低表达组,使用survminer package [0.4.9]绘制和可视化OS曲线,同时探讨年龄、性别等亚组对胰腺癌预后的意义。
通过UCSC XENA RNAseq 提取胰腺癌基因组,在GTEx数据库提取相应的正常组织基因进行分析。利用ROCpackage [1.17.0.1]和ggplot2 package [3.3.3]绘制和可视化PLAU的和ROC曲线。
通过R语言利用survival package [3.2-10] (用于生存数据的统计分析)对胰腺癌基因组图谱进行分析,通过RNAseq数据对纳入变量进行COX回归分析。
2.4. 免疫浸润分析
利用R GSVA package [1.34.0],通过ssGSEA算法分析PLAU在胰腺癌各种免疫细胞(包括aDC;B细胞;CD8 T细胞;细胞毒性细胞;DC;嗜酸性粒细胞;iDC;巨噬细胞;肥大细胞;中性粒细胞;NK cd56bright;NK cd56dim;NK细胞;pDC;T细胞;T辅助细胞;Tcm、Tem、Tfh、Tgd、Th1、Th17、Th2、Treg细胞)中的表达程度和P值。
2.5. 实时定量PCR
从Panc-1、hTERT-HPNE和缺氧培养24 h后的panc-1细胞中提取总RNA。使用PrimeScrip RT-PCR试剂盒(Takara,日本)进行反转录,使用SYBR PreMix Ex Taq (Takara,日本)在罗氏仪器上进行qRT-PCR。本研究使用的引物序列如下:PLAU上游引物:GGGAATGGTCACTTTTACCGAG,下游引物: GGGCATGGTACGTTTGCTG;GAPDH上游引物:GGAGCGAGATCCCTCCAAAAT,下游引物: GGCTGTTGTCATACTTCTCATGG。
2.6. 统计分析
统计分析使用SPSS 24.0软件(IBM Corp, NY, USA),图像处理使用GraphPad Prism 8 (GraphPad, USA)。t检验分析基因差异。以|logFC| > 1和P < 0.05为阈值选择显著性差异表达基因。
3. 结果
3.1. 基因表达及富集分析
共筛选出9222个差异表达基因,其中上调基因2456个,下调基因158个(图1(A))。
通过GO/KEGG分析这2614个中与缺氧途径相关的基因。确定了缺氧的通路:GO:0001666 (对缺氧的反应,共鉴定出66个基因) (图1(B))。
选择PLAU作为关键基因,探讨其是否在胰腺癌缺氧和免疫浸润中起重要作用。
为了研究PLAU是否在PAAD中起作用,采用RNAseq基因表达谱分析比较PLAU高表达组和低表达组的基因表达谱,用柱状图显示PLAU在正常组织和肿瘤组织中的表达,结果显示有显著差异(图1(C))。
我们还发现除了在胰腺癌中,PLAU在肾上腺皮质癌、乳腺癌、宫颈鳞状细胞癌、胆管癌、结肠癌、弥漫性大B细胞淋巴瘤、食管癌、多形性胶质母细胞瘤、头部和颈部鳞状细胞癌、低级别神经胶质瘤等肿瘤中均呈高表达;PLAU在腺样囊性癌、肾嫌色细胞癌、肾透明细胞癌、肾乳头状细胞癌等肿瘤中呈低表达(图1(D))。
通过HPA免疫组化染色图像,我们发现PLAU在胰腺癌组织中的表达(图1(E))高于正常胰腺组织(图1(F))。

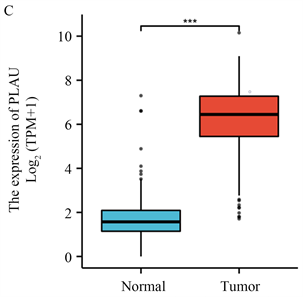
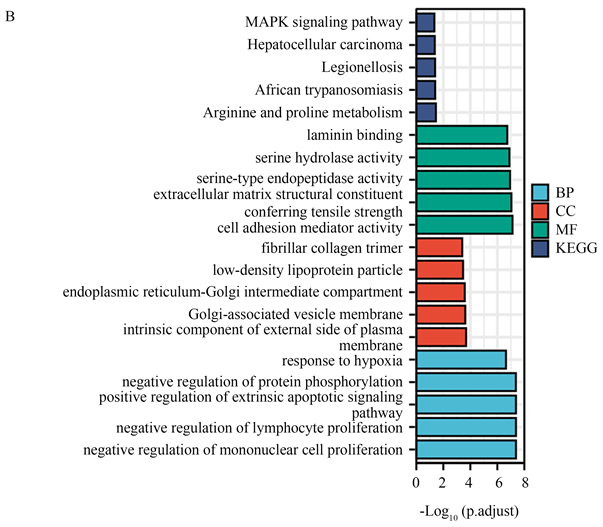
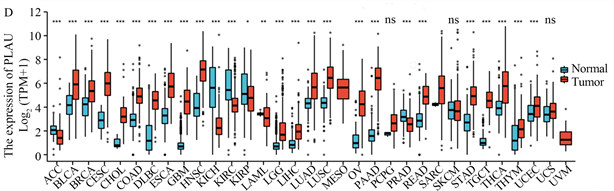
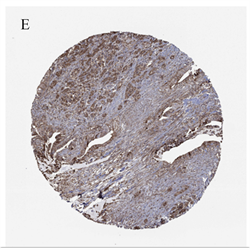 (A) 来自GEPIA2的胰腺癌差异基因,红色为上调,蓝色为下调;(B) 通过GO/KEGG筛选来自GEPIA2胰腺癌的差异基因;(C) PLAU在胰腺癌和正常胰腺组织中的表达;(D) PLAU在泛癌中的表达;(E) PLAU在胰腺癌组织中免疫组化染色图像;(F) PLAU在正常组织中免疫组化染色图像。
(A) 来自GEPIA2的胰腺癌差异基因,红色为上调,蓝色为下调;(B) 通过GO/KEGG筛选来自GEPIA2胰腺癌的差异基因;(C) PLAU在胰腺癌和正常胰腺组织中的表达;(D) PLAU在泛癌中的表达;(E) PLAU在胰腺癌组织中免疫组化染色图像;(F) PLAU在正常组织中免疫组化染色图像。
Figure 1. Gene expression and enrichment analysis
图1. 基因表达及富集分析
3.2. 生存与预后分析
根据OS曲线,我们发现PLAU高表达患者的生存时间相较于PLAU低表达患者的生存时间明显缩短(图2(A))。胰腺癌PLAU的ROC曲线显示其AUC为0.976,诊断效果显著(图2(B))。
根据研究,我们发现PLAU高表达患者在T2/T3期(图2(C))、男性(图2(D))和65岁以下亚组(图2(E))的生存时间分布有统计学差异。
单因素COX回归发现PLAU、N1期和病理期II期是胰腺癌的独立预后因素。多因素cox回归显示,只有PLAU和N1期是胰腺癌的独立预后因素(表1)。
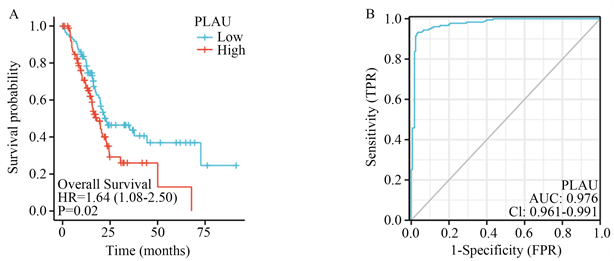
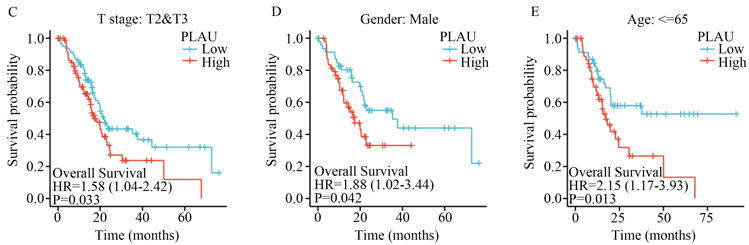 (A) PLAU高表达患者和PLAU低表达患者的os曲线;(B) 胰腺癌中PLAU的ROC曲线;(C) PLAU表达与T2/T3期胰腺癌患者生存时间;(D) PLAU表达与的男性胰腺癌患者生存时间;(E) PLAU表达与65岁以下亚组的生存时间。
(A) PLAU高表达患者和PLAU低表达患者的os曲线;(B) 胰腺癌中PLAU的ROC曲线;(C) PLAU表达与T2/T3期胰腺癌患者生存时间;(D) PLAU表达与的男性胰腺癌患者生存时间;(E) PLAU表达与65岁以下亚组的生存时间。
Figure 2. Survival and prognosis analysis of PLAU expression
图2. PLAU表达的生存与预后分析
Table 1. Univariate and multivariate COX regression analysis of pancreatic cancer
表1. 胰腺癌的单因素和多因素COX回归分析
3.3. PLAU表达与免疫浸润的相关性
我们发现PLAU与胰腺癌中锚定依赖细胞、树突状细胞、巨噬细胞、中性粒细胞、NK CD56dim细胞、NK细胞、辅助性T细胞、Tem、Tgd、Th1、Th2细胞和Treg细胞的浸润程度呈正相关。与Th17细胞浸润程度呈负相关,具有显著性(图3(A))。同时,我们还发现PLAU高表达组与低表达组相比,NK细胞、NK CD56dim细胞、中性粒细胞、巨噬细胞、iDC、Th1细胞、Th17细胞、Th2细胞和Treg的分布有所不同(图3(B))。
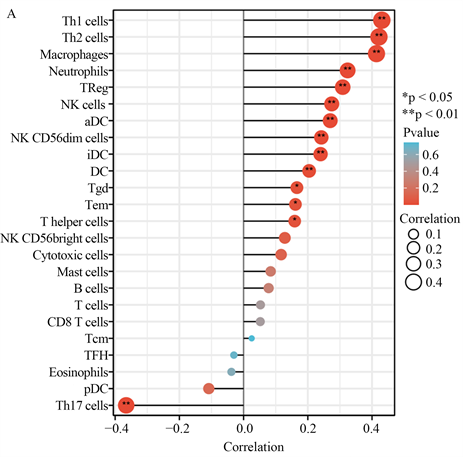
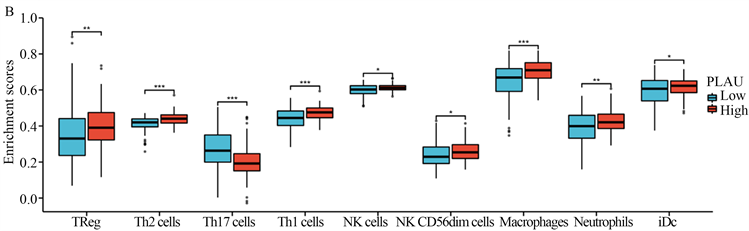 (A) PLAU表达与胰腺癌免疫细胞浸润程度的相关性;(B) PLAU表达与胰腺癌免疫细胞浸润分布的相关性。
(A) PLAU表达与胰腺癌免疫细胞浸润程度的相关性;(B) PLAU表达与胰腺癌免疫细胞浸润分布的相关性。
Figure 3. Correlation between PLAU expression and immune infiltration
图3. PLAU表达与免疫浸润的相关性分析
3.4. 实时定量PCR
根据RT-PCR结果,我们发现PLAU在panc-1细胞中较hTERT-HPNE细胞表达增高(图4(A))。缺氧处理的panc-1细胞表达低于正常培养的panc-1细胞(图4(B))。
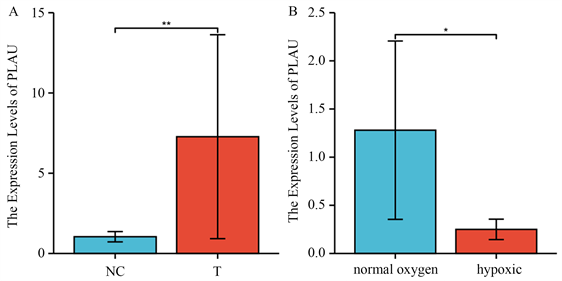 (A) PLAU在Panc-1细胞和正常人胰管细胞中的表达;(B) PLAU在正常培养的Panc-1细胞和缺氧Panc-1细胞中的表达。
(A) PLAU在Panc-1细胞和正常人胰管细胞中的表达;(B) PLAU在正常培养的Panc-1细胞和缺氧Panc-1细胞中的表达。
Figure 4. Differential expression of PLAU in panc-1, normal human pancreatic duct and hypoxic Panc-1 cells
图4. PLAU在Panc-1、正常人胰管和缺氧Panc-1细胞中的表达差异
4. 讨论
胰腺癌是目前世界上恶性程度、致死率最高的恶性肿瘤之一 [25]。现如今多数癌症的存活率较前已经显著提高,但胰腺癌却没有 [26]。多数胰腺癌一旦发现即进入中晚期,其对大多数治疗方案无效,导致总体预后差 [7]。因此,这就需要找到一种可靠的预后标志物。在我们的研究中,我们通过搜索GEO、TCGA等在线数据库,获得可靠的RNAseq数据和相应的临床数据,研究PLAU在胰腺癌中的表达和预后,结果显示,PLAU在胰腺癌组织中的表达明显高于正常组织。同时,我们发现PLAU在多种癌症中的表达存在显著差异。PLAU高表达的胰腺癌患者预后较PLAU低表达的患者差,生存时间在T2/T3期患者、男性患者、小于65岁患者中的分布也有显著差异。同时,通过单因素/多因素COX回归分析,我们发现PAAD中PLAU的表达与肿瘤T分期和N分期相关。提示PLAU可能是预测PAAD预后的生物标志物和治疗靶点。
根据GO/KEGG分析,我们发现PLAU出现在三个低氧通路中,根据RT-qPCR的结果,我们发现胰腺癌细胞PANC-1经缺氧处理后其PLAU的表达低于正常胰腺癌细胞,这说明缺氧是影响PLAU表达的因素之一。而PLAU是胰腺癌可靠的预后分子之一,缺氧可能通过影响PLAU的表达来影响胰腺癌的预后。
肿瘤的进展和预后与免疫细胞在肿瘤微环境中的浸润密切相关 [27]。PLAU的表达受胰腺癌不同的免疫浸润水平的影响,PLAU的表达也影响免疫浸润的程度,这表明PLAU可以调节胰腺癌的免疫微环境。Treg细胞可降低CD4+,CD8+ NK细胞的活性被认为是已知的最有效的肿瘤免疫抑制细胞 [28]。Th2通过释放IL-4和IL-13促进肿瘤发生,并通过介导凋亡抵抗和增强代谢促进增殖、迁移和侵袭 [29]。同时,PLAU与Th17细胞在胰腺癌中的分布呈负相关。Th17细胞是一种促进免疫功能的细胞,PLAU在胰腺癌中的高表达可以抑制Th17细胞的分布,维持胰腺癌免疫抑制的肿瘤微环境 [30]。我们发现PLAU表达与TH2和Treg细胞水平呈正相关,与Th17细胞水平呈负相关,提示在胰腺癌中过表达PLAU可以激活这两类免疫抑制细胞,抑制免疫激活细胞,有助于维持胰腺癌的免疫抑制微环境。
PLAU在胰腺癌细胞中的差异表达提示其是一种关键的预后基因。它与胰腺癌的缺氧和免疫侵袭有关,在胰腺癌的肿瘤微环境中起重要作用。这些发现为研究胰腺癌的分子机制和新的治疗靶点提供了新的方向。
基金项目
国家自然科学基金委员会资助面上项目(No. 52075277)。
文章引用
刘任政,吴仰东,孙 琳,胡 骁. 缺氧和免疫抑制的胰腺癌肿瘤微环境中PLAU的表达及对预后的影响
Expression of PLAU in Tumor Microenvironment of Hypoxic and Immunosuppressive Pancreatic Cancer and Its Influence on Prognosis[J]. 临床医学进展, 2023, 13(01): 684-694. https://doi.org/10.12677/ACM.2023.131100
参考文献
- 1. Midha, S., Chawla, S., and Garg, P.K. (2016) Modifiable and Non-Modifiable Risk Factors for Pancreatic Cancer: A Review. Cancer Letters, 381, 269-277. https://doi.org/10.1016/j.canlet.2016.07.022
- 2. Huang, S., Zhao, J.L., Song, J.X., et al. (2021) Interferon Alpha-Inducible Protein 27 (IFI27) Is a Prognostic Marker for Pancreatic Cancer Based on Comprehensive Bioinformatics Analysis. Bioengineered, 12, 8515-8528. https://doi.org/10.1080/21655979.2021.1985858
- 3. Cao, F., Li, J., Li, A. and Li, F. (2017) Radical Antegrade Modular Pancreatosplenectomy versus Standard Procedure in the Treatment of Left-Sided Pancreatic Cancer: A Systemic Review and Meta-Analysis. BMC Surgery, 17, Article No. 67. https://doi.org/10.1186/s12893-017-0259-1
- 4. Koller, M., Hartmans, E., de Groot, D.J.A., et al. (2017) Da-ta-Driven Prioritization and Review of Targets for Molecular-Based Theranostic Approaches in Pancreatic Cancer. Jour-nal of Nuclear Medicine, 58, 1899-1903. https://doi.org/10.2967/jnumed.117.198440
- 5. Ansari, D., Tingstedt, B., Andersson, B., et al. (2016) Pancreatic Cancer: Yesterday, Today and Tomorrow. Future Oncology, 12, 1929-1946. https://doi.org/10.2217/fon-2016-0010
- 6. Marschner, N., Hegewisch-Becker, S., Reiser, M., et al. (2023) FOLFIRINOX or Gemcitabine/Nab-Paclitaxel in Advanced Pancreatic Adenocarcinoma: A Novel Validated Prognostic Score to Facilitate Treatment Decision-Making in Real-World. International Journal of Cancer, 152, 458-469. https://doi.org/10.1002/ijc.34271
- 7. Rouanet, M., Lebrin, M., Gross, F., et al. (2017) Gene Therapy for Pancre-atic Cancer: Specificity, Issues and Hopes. International Journal of Molecular Sciences, 18, Article No. 1231. https://doi.org/10.3390/ijms18061231
- 8. Grossberg, A.J., Chu, L.C., Deig, C.R., et al. (2020) Multidisciplinary Standards of Care and Recent Progress in Pancreatic Ductal Adenocarcinoma. CA: A Cancer Journal for Clinicians, 70, 375-403. https://doi.org/10.3322/caac.21626
- 9. Dougan, S.K. (2017) The Pancreatic Cancer Microenvironment. The Cancer Journal, 23, 321-325. https://doi.org/10.1097/PPO.0000000000000288
- 10. Morrison, A.H., Byrne, K.T. and Vonderheide, R.H. (2018) Immunotherapy and Prevention of Pancreatic Cancer. Trends in Cancer, 4, 418-428. https://doi.org/10.1016/j.trecan.2018.04.001
- 11. Huber, M., Brehm, C.U., Gress, T.M., et al. (2020) The Immune Microenvironment in Pancreatic Cancer. International Journal of Molecular Sciences, 21, Article No. 7307. https://doi.org/10.3390/ijms21197307
- 12. Schwartz, D.L., Bankson, J.A., Lemos, R., et al. (2010) Radiosensitiza-tion and Stromal Imaging Response Correlates for the HIF-1 Inhibitor PX-478 Given with or without Chemotherapy in Pancreatic Cancer. Molecular Cancer Therapeutics, 9, 2057-2067. https://doi.org/10.1158/1535-7163.MCT-09-0768
- 13. Huang, C.H., Xiang, Y.K., Chen, S.C., et al. (2017) Demo-kine Contributes to Epithelial-Mesenchymal Transition through Increased Activation of Signal Transducer and Activator of Transcription 3 in Pancreatic Cancer. Cancer Science, 108, 2130-2141. https://doi.org/10.1111/cas.13347
- 14. Minchenko, O.H., Tsuchihara, K., Minchenko, D.O., Bikfalvi, A. and Esumi, H. (2014) Mechanisms of Regulation of PFKFB Expression in Pancreatic and Gastric Cancer Cells. World Jour-nal of Gastroenterology, 20, 13705-13717. https://doi.org/10.3748/wjg.v20.i38.13705
- 15. Carstens, J.L., Correa de Sampaio, P., Yang, D., et al. (2017) Spa-tial Computation of Intratumoral T Cells Correlates with Survival of Patients with Pancreatic Cancer. Nature Communi-cations, 8, Article No. 15095. https://doi.org/10.1038/ncomms15095
- 16. Lei, X., Lei, Y., Li, J.K., et al. (2020) Immune Cells within the Tumor Microenvironment: Biological Functions and Roles in Cancer Immunotherapy. Cancer Letters, 470, 126-133. https://doi.org/10.1016/j.canlet.2019.11.009
- 17. Duewell, P., Steger, A., Lohr, H., et al. (2014) RIG-I-Like Hel-icases Induce Immunogenic Cell Death of Pancreatic Cancer Cells and Sensitize Tumors toward Killing by CD8+ T Cells. Cell Death & Differentiation, 21, 1825-1837. https://doi.org/10.1038/cdd.2014.96
- 18. Feng, M.Y., Xiong, G.B., Cao, Z., et al. (2017) PD-1/PD-L1 and Immu-notherapy for Pancreatic Cancer. Cancer Letters, 407, 57-65. https://doi.org/10.1016/j.canlet.2017.08.006
- 19. Ai, C., Zhang, J.X., Lian, S.Y., et al. (2020) FOXM1 Functions Collaboratively with PLAU to Promote Gastric Cancer Progression. Journal of Cancer, 11, 788-794. https://doi.org/10.7150/jca.37323
- 20. Lin, M., Zhang, Z., Gao, M., et al. (2019) MicroRNA-193a-3p Suppresses the Colorectal Cancer Cell Proliferation and Progression through Down-regulating the PLAU Expression. Cancer Management and Research, 11, 5353-5363. https://doi.org/10.2147/CMAR.S208233
- 21. Li, Y., Lu, Z.L., Che, Y., et al. (2017) Immune Signature Profiling Identified Predictive and Prognostic Factors for Esophageal Squamous Cell Carcinoma. Oncoimmunology, 6, e1356147. https://doi.org/10.1080/2162402X.2017.1356147
- 22. Moirangthem, A., Bondhopadhyay, B., Mukherjee, M., et al. (2016) Simultaneous Knockdown of uPA and MMP9 Can Reduce Breast Cancer Progression by Increasing Cell-Cell Adhesion and Modulating EMT Genes. Scientific Reports, 6, Article No. 21903. https://doi.org/10.1038/srep21903
- 23. Mauro, C.D., Pesapane, A., Formisano, L., et al. (2017) Urokinase-Type Plasminogen Activator Receptor (uPAR) Expression Enhances Invasion and Metastasis in RAS Mutated Tumors. Scien-tific Reports, 7, Article No. 9388. https://doi.org/10.1038/s41598-017-10062-1
- 24. Tang, Z.F., Li, C.W., Kang, B.X., et al. (2017) GEPIA: A Web Server for Cancer and Normal Gene Expression Profiling and Interactive Analyses. Nucleic Acids Research, 45, W98-W102. https://doi.org/10.1093/nar/gkx247
- 25. Vincent, A., Herman, J., Schulick, R., Hruban, R. and Gog-gins, M. (2011) Pancreatic Cancer. Lancet, 378, 607-620. https://doi.org/10.1016/S0140-6736(10)62307-0
- 26. Collisson, E.A., Bailey, P., Chang, D.K. and Biankin, A.V. (2019) Molecular Subtypes of Pancreatic Cancer. Nature Reviews Gastroenterology & Hepatology, 16, 207-220. https://doi.org/10.1038/s41575-019-0109-y
- 27. Roth, S., Zamzow, K., Gaida, M.M., et al. (2020) Evolution of the Immune Landscape during Progression of Pancreatic Intraductal Papillary Mucinous Neoplasms to Invasive Cancer. eBioMedicine, 54, Article ID: 102714. https://doi.org/10.1016/j.ebiom.2020.102714
- 28. Padoan, A., Plebani, M., and Basso, D. (2019) Inflammation and Pancreatic Cancer: Focus on Metabolism, Cytokines, and Immunity. International Journal of Molecular Sciences, 20, Article No. 676. https://doi.org/10.3390/ijms20030676
- 29. Shi, J.W., Song, X.J., Traub, B., et al. (2021) In-volvement of IL-4, IL-13 and Their Receptors in Pancreatic Cancer. International Journal of Molecular Sciences, 22, Ar-ticle No. 2998. https://doi.org/10.3390/ijms22062998
- 30. Lee, G.R. (2018) The Balance of Th17 versus Treg Cells in Autoimmunity. International Journal of Molecular Sciences, 19, Article No. 730. https://doi.org/10.3390/ijms19030730
NOTES
*通讯作者Email: huxiao202@163.com
