Pure Mathematics
Vol.
13
No.
05
(
2023
), Article ID:
66011
,
14
pages
10.12677/PM.2023.135130
基于网络方法确定乳腺癌转移相关基因
田智萌,张子豪
山东科技大学,数学与系统科学学院,山东 青岛
收稿日期:2023年4月12日;录用日期:2023年5月15日;发布日期:2023年5月22日

摘要
乳腺癌是女性最常见的恶性肿瘤之一,其发展是一个长期的、多阶段、多基因改变积累的过程。临床及实验研究发现,在乳腺癌发展与转移的过程中,许多原癌基因、抑癌基因及相关蛋白质发生改变。本文提出了一种结合随机游走重启算法的综合模型,并将其应用于乳腺癌骨转移关键候选基因的鉴定中,在蛋白质相互作用网络上识别乳腺癌转移候选基因。首先利用RWR算法和置换检验规则对基因进行预选,得到候选基因集。然后构建一个候选基因间的关键子网并计算关键子网中节点的介数中心性。最后,利用交互得分规则对基因筛选,选择介数得分排名前三十的基因作为乳腺癌骨转移关键基因。相互作用分析、富集分析及文献挖掘的结果表明26个潜在的关键基因都参与了乳腺癌骨转移的起始或进程,验证了该综合模型的有效性。
关键词
肿瘤转移,复杂网络,蛋白质相互作用,随机游走重启算法,介数中心性

Network-Based Method for the Identification of Genes Related to Cancer Metastasis
Zhimeng Tian, Zihao Zhang
College of Mathematics and System Science, Shandong University of Science and Technology, Qingdao Shandong
Received: Apr. 12th, 2023; accepted: May 15th, 2023; published: May 22nd, 2023

ABSTRACT
Breast cancer is one of the most common malignant tumors in women, and its development is a long-term, multi-stage, multi-genetic alteration accumulation process. Clinical and experimental studies have revealed that many proto-oncogenes, oncogenes and related proteins are altered during the process of breast cancer development and metastasis. In this paper, we propose an integrated model combining random wander restart algorithm and apply it to the identification of key candidate genes for breast cancer bone metastasis on protein interaction network to identify breast cancer metastasis candidate genes. The genes are first pre-selected using the RWR algorithm and the substitution test rule to obtain the candidate gene set. Then a key subnet among candidate genes is constructed and the betweenness centrality of nodes in the key subnet is calculated. Finally, the genes were screened using the interaction score rule, and the top thirty genes in the mediator score were selected as the key genes for breast cancer bone metastasis. The results of interaction analysis, enrichment analysis and literature mining indicated that 26 potential key genes were involved in the initiation or progression of breast cancer bone metastasis, validating the validity of this comprehensive model.
Keywords:Tumor Metastasis, Complex Networks, Protein Interactions, RWR, Betweenness Centrality

Copyright © 2023 by author(s) and Hans Publishers Inc.
This work is licensed under the Creative Commons Attribution International License (CC BY 4.0).
http://creativecommons.org/licenses/by/4.0/


1. 引言
乳腺癌是女性最常见的恶性肿瘤之一,其发展是一个长期的、多阶段、多基因改变积累的过程 [1] [2] ,病因与发病机制一直是乳腺癌研究领域的重点。乳腺肿瘤的发展过程一般可分为以下三个阶段:起始、侵袭和转移。在乳腺癌中,常见的转移部位包括骨头、肝脏和大脑等,并且它们有完全不同的症状。其中,超过70%的乳腺癌患者死亡可归因于乳腺癌骨转移 [3] [4] 。除此之外,骨转移还同时伴随不明原因的体重下降、骨头关节疼痛、黄胆及神经症状等。因此,本研究以乳腺癌骨转移作为研究案例具有特殊的意义和价值。临床及实验研究发现,在乳腺癌发展与转移的过程中,许多原癌基因、抑癌基因及相关蛋白质发生改变 [5] 。
2011年,Ruan等人 [6] 整合蛋白质相互作用网络和基因表达数据,提出一种鉴定乳腺癌转移中的生物标志物的方法。该方法应用于两个乳腺癌微阵列数据集的生物标志物的发现,这些生物标记物中含有大量已知的癌症易感基因,并在致癌过程中涉及的生物学过程和通路中显著富集。此外,该方法选择的标记在两个数据集中具有比以往研究更高的稳定性。2016年,Tian等人 [7] 提出了一种结合蛋白质相互作用信息与表达数据的方法来检索乳腺癌转移相关基因。该论文基于随机森林MDG准则和蛋白质相互作用信息的GeneRank算法两种变量选择算法的重要得分,提出了一种新的基于随机森林的算法,来识别与乳腺癌转移高度相关的重要变量。较高的预测精度充分说明了通过该算法选择基因表的可靠性和可解释性。2017年,Cai等人 [8] 提出了一种基于蛋白质相互作用网络使用最短路径算法识别乳腺癌骨转移关键基因的方法。该方法在已确认的乳腺癌和骨癌相关基因数据的基础上,将最短路径算法应用于蛋白质相互作用网络,最终得到18个介导乳腺癌骨转移的候选基因,并进行了广泛的分析,根据研究证实了这些基因参与了乳腺癌骨转移过程。
本文在第一部分介绍了当前国内外的乳腺癌转移研究,在第二部分介绍了数据来源并提出了一种结合随机游走重启算法和介数中心性的综合模型,在第三部分将模型应用于乳腺癌骨转移关键候选基因的识别,在第四部分通过相互作用分析、富集分析及文献挖掘的分析结果从基因层面揭示了肿瘤转移机制。
2. 基于随机游走重启算法的乳腺癌转移关键基因识别
2.1. 数据与数据预处理
乳腺癌相关基因主要来源于Oncomine数据库(https://www.oncomine.org)和TCGA数据库(https://www.cancer.gov/ccg/rresearc/tcga)。Oncomine数据库是一个癌症微阵列数据库,提供了多种癌症类型的转录组数据。通过对两个数据库的检索,并对得到的基因进行整合,将它们映射到STRING数据库中的Ensembl ID上。最终得到了358个乳腺癌相关基因,其集合用 表示;得到的588个骨癌相关基因,其集合用 表示。
2.2. 方法介绍
在构建好的蛋白质相互作用网络上,本研究提出了一种基于随机游走重启算法的癌症转移基因识别方法,并将其运用于乳腺癌骨转移特异性关键基因的鉴定中。整个模型的流程见图1所示。
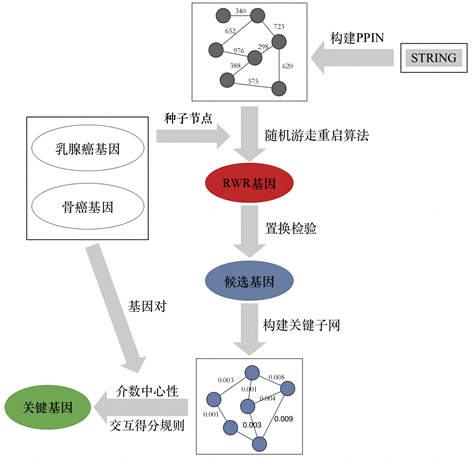
Figure 1. The flow chart of our model
图1. 整个模型的流程图
2.2.1. 随机游走重启算法
RWR算法是一种经典的排序算法,它从一些种子节点开始,模拟其在网络中随机游走和重启,同时更新所有节点的概率得分并对节点进行排名。在本方法中,将已验证的乳腺癌与骨癌的基因作为种子节点,在PPIN上执行RWR算法,利用算法的结果对网络中其它节点进行排名,以实现对基因的预选,其主要过程如下:
其中,重启概率 表示在随机游走过程中回到种子节点的概率,取值范围为 。当 时,种子节点的信息对于结果无影响,节点的概率得分排名由节点在网络结构中的位置决定。根据先前结合RWR算法应用于基因识别的研究,当 时,算法的结果可以更好地融合种子节点的信息和网络的结构的影响。因此,本研究将重启概率 设为0.8来表示种子节点的重要性。当 ,迭代结果趋于稳定,算法停止并输出 。在 中,每个节点(基因)的概率得分表示其与种子节点在PPIN中的相似性得分。因此,具有较高概率得分的基因与已验证的骨癌和乳腺癌基因更相关,从而更有可能是转移相关基因。为了防止丢失潜在的转移相关基因,将概率得分的阈值设为 ,即节点概率得分大于 对应的基因被筛选出来,这些基因被记为RWR基因。与其他算法筛选相关基因相比,RWR具有算法快速、简单及准确性高等特点。
2.2.2. 置换检验规则
上述RWR基因可能会受到PPIN结构的影响,从而存在很多与癌症转移无关的假阳性基因。例如,在网络中度中心性远高于平均度的节点即使与种子节点无关,也会获得较大的概率得分,因此使用随机选择网络中的某些节点作为种子节点这些基因仍会得到较高的概率得分排名。基于这一思想,本研究提出利用置换检验规则来筛选并删除这些假阳性基因。
首先,将总置换数(total permutations)设为1000,即随机构建了1000个Ensembl IDs集合,记为 ,每个集合包含882个随机的基因Ensembl IDs。然后,通过将 中的882个基因设置为种子节点,在PPIN上执行RWR算法以获取每个RWR基因的概率得分。对于每个RWR基因,存在一个真实概率得分 和1000个随机概率得分 。最后,对每一个RWR基因 ,计算p-value值如下:
(2-2)
其中, 表示1000个随机概率得分 中大于真实概率得分 的个数,如果随机概率得分 普遍大于真实概率得分 ,说明 更可能是因为网络结构而被选出来的假阳性基因。显然,p-value值很高的RWR基因应当被删除。由于0.05是作为被广泛接受的统计学检验传统显著性水平的阈值,本研究选择p-value值小于0.05的RWR基因作为乳腺癌骨转移的潜在候选基因做进一步分析。
2.2.3. 构建关键子网
经过RWR算法和置换检验得到的可能仅与乳腺癌基因或骨癌基因中一项高度相关,为了从候选基因中筛选出在转移过程中起到更关键作用的基因,本研究从PPIN中除候选基因对应节点以外的所有节点及连边删除,得到了一个仅保留候选基因间相互作用的关键子网。
在PPIN中,蛋白质之间连边的权重代表蛋白质间的相互作用得分,其取值范围为 。在基于最短路径的模型上,节点之间通常使用较低的权重来表示更强的相关性。因此,对于关键子网中蛋白质 和 ,当相互作用得分 时,利用下面的公式更新它们连边的权重。
(2-3)
显然,当权重值 越小,表示关键子网中蛋白质之间的相互作用更强。根据权重更新公式函数图像,见图2可知,当相互作用得分 增大时,函数曲线的斜率减小,更新后的权重相差更小。因此,通过公式(2-3)对关键子网的权重进行归一化处理后,权重的分布相对之前更加集中。
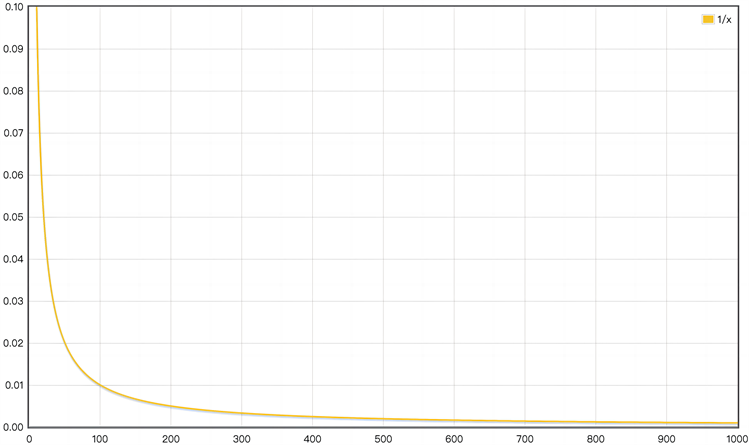
Figure 2. Image of weight update formula
图2. 权重更新公式函数图像
2.2.4. 介数中心性
通过删除网络中的无关基因并更新节点间的权重,得到了一个候选基因间的关键子网。本研究中,与乳腺癌和骨癌相关基因都有密切关联的基因更有可能是与转移有关的特异性基因。在该关键子网中,我们应用dijkstra算法计算乳腺癌关键基因集 中所有基因到骨癌关键基因集 中所有基因的最短路径。在每条最短路径上,连续的节点通过较低权重的边相连,表示了相对应蛋白质之间的强相关性。由于每条最短路径的起始和末端节点为已验证的乳腺癌和骨癌相关基因,这些最短路径内部节点所对应的基因可能是导致乳腺癌向骨转移的特异性基因。
对于除去882个种子节点对应基因以外的每个候选基因 ,计算它基于所有基因对 的介数中心性得分:
(2-4)
其中 代表在所有最短路径中包含基因 对应节点的数目; 指 和 中基因数目的乘积,表示可能存在的所有最短路径数目。介数中心性得分 表示了最短路径经过基因 的概率,因此 值更大的候选基因更有可能是与乳腺癌骨转移有关的关键基因。
此外,基于PPI中相互作用得分较高的两个蛋白质更有可能具有相似功能的研究,利用公式(2-5)计算了每个候选基因 的最大–最小交互得分MMIS。
(2-5)
本研究以STRING数据库中最高置信度值900作为MMIS的阈值,最终选择了MMIS不小于900且介数中心性得分BC排名前三十的基因作为乳腺癌骨转移的潜在关键基因,这些基因被认为在乳腺癌骨转移中发挥了重要作用。
3. 结果与分析
通过将已验证的882个乳腺癌与骨癌相关基因作为种子节点,在PPIN上执行RWR算法,选择结果中概率得分大于 的基因,得到了7105个RWR基因。此外,为了消除网络结构的假阳性影响,本研究进行了置换检验规则并选择p-value值小于0.1的2955个基因作为乳腺癌骨转移的候选基因。
为了筛选在乳腺癌骨转移过程中起关键作用的基因,本研究构建了一个候选基因间的关键子网,并在该网络上应用最短路径算法。然后,计算了每个候选基因的最大–最小交互得分MMIS和介数中心性得分BC,最后选择了MMIS不小于900且介数中心性得分BC排名前三十的基因作为乳腺癌骨转移的潜在关键基因,它们的Ensembl ID、基因名、P-value值、MMIS值及验证的参考文献在表1中展示。
Table 1. Thirty potentially core genes identified in our method and ranked by the BC value
表1. 通过该方法得到的30个潜在的关键基因根据BC的排名
4. 分析与讨论
根据文献中的实验证据,通过该方法得到的30个潜在关键基因中,有26个基因已被证实与乳腺癌的骨转移中的基因组整合、转移的发展过程或特定并发症有关。这些发现证明了本研究所提出的综合方法的有效性和合理性。
4.1. 相互作用分析
为了验证这些关键基因与已验证的乳腺癌和骨癌相关基因之间的关系,将这30个潜在的关键基因和集合 、 中重复的50个基因输入STRING数据库中,得到了它们之间的相互作用网络如图3所示。本研究将网络中边权重的阈值设为700,仅当节点对应蛋白质之间的相互作用得分大于或等于700时,它们之间的连边在网络中保留。图中左边的26个节点为已被证实与乳腺癌的骨转移有关的潜在关键基因,中间的节点表示已验证的乳腺癌和骨癌相关基因,右边的四个节点表示还未被实验证明的潜在关键基因。该蛋白质网络的节点平均度为13.5,相对于一般的网络节点间联系更紧密,表示蛋白质之间的强相关性。其中,26个已被证实与乳腺癌的骨转移有关的潜在关键基因对应节点的平均度为20.6,未验证的4个基因的平均节点度为22.8,暗示了这些潜在关键基因在乳腺癌骨转移中的重要作用。
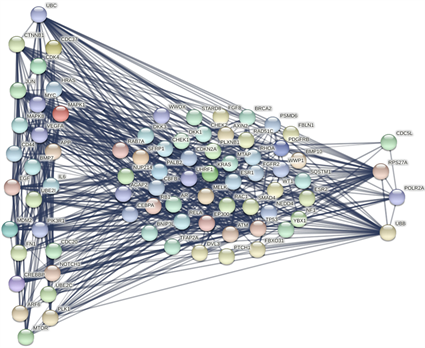
Figure 3. The interactions between known both breast and bone cancer-related genes and the thirty potential core genes
图3. 已知的乳腺癌和骨癌相关基因与30个关键基因之间的相互作用图
4.2. 基因富集分析
为了揭示这30个潜在关键基因的分子功能及生物学背景,本研究使用了DAVID功能注释工具。DAVID是一个功能强大的注释工具,可用于对大型基因列表进行整合和系统化,并根据KEGG通路和GOterms对基因进行分析。其中GOterms主要包含三个部分:生物过程(BP),细胞成分(CC)和分子功能(MF)。由于本研究提出的方法主要是通过蛋白质间相互作用来鉴定基因,因此基于BP进行分析更为合理。
在基于BP的富集分析中,30个潜在关键基因显著富集在15个GOterms上,图4展示了十个主要的GOterms名称及每个GOterms富集的基因数量。文献综述显示,大多数富集的GOterms与乳腺癌骨转移的生物过程有关。例如,GO:0045944-RNA聚合酶II启动子转录的正调控富集了包括AP1、HARS和CREBBP等14个基因,它主要激活了RNA聚合酶II启动子转录过程的频率、速率及程度,并与骨癌的形成和转移相关 [6] [58] 。GO:0038128-ERBB2信号通路的P-value值最低,该信号通路已被研究证明在乳腺癌的骨转移过程中被激活 [7] 。此外,GO:0008284-细胞增殖的正向调控和GO:0007219-Notch信号通路被报道促进了骨肿瘤细胞的形成 [8] [59] 。
在基于KEGG通路的分析中,30个潜在关键基因显著富集在14条通路上,图5展示了十个主要的KEGG通路名称及每条通路富集的基因数量。其中,hsa05200-癌症通路是富集最显著的通路,包含UBC、AP1、VEGF和FN1等15个基因。该通路富集的基因通常参与癌症的形成和发展,从而体现了该方法的有效性。hsa04151-PI3K-Akt信号通路富集了包括CDC37、MDM2和PIK3R1等12个基因,它是一种在调节代谢、细胞生长和血管生成中发挥重要作用的细胞内信号转导通路,已证明与乳腺癌的骨转移高度相关 [60] [61] 。此外,hsa04012-ErbB信号通路、hsa04350-TGF-β信号通路等KEGG通路已被证明有助于乳腺癌的骨转移过程 [62] [63] [64] 。
富集分析的结果表明,这些基因的功能活性和定位与乳腺癌骨转移过程直接或间接相关,进一步支持了提出方法的结果。此外,这些发现可能会拓展探索乳腺癌骨转移新机制的途径。
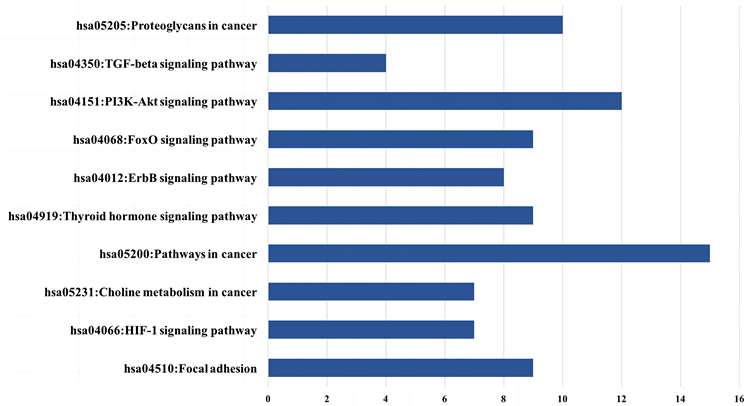
Figure 4. Ten crucial GO terms enriched by potential core genes
图4. 潜在关键基因富集的十个主要GO terms
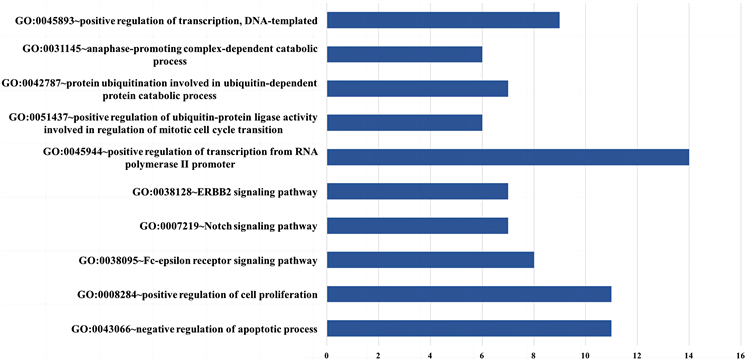
Figure 5. Ten crucial KEGG pathways enriched by 30 potential core genes
图5. 潜在关键基因富集的十个主要KEGG通路
4.3. 基因与转移机制分析
在30个潜在的关键基因中,有26个基因已通过文献中的实验证据证实与乳腺癌骨转移的形成或发展有关。在所有关键基因中,介数中心性BC值排名最高的基因UBC (泛素C)已被证实参与了乳腺癌的转移过程。体外实验证明,UBC通过调节控制细胞周期和增殖的中枢基因在骨髓间充质干细胞的复制性衰老中起着关键作用,这一过程促进了骨骼组织中乳腺癌细胞的适应性和侵袭性 [9] 。此外,UBC末端水解酶-L3促进了EMT过程,这是癌细胞侵袭和转移的重要因素 [10] 。关键基因中另一个与泛素有关的基因UBE2I参与编码酵母泛素结合酶的人类同源物,对调节细胞周期至关重要。研究表明,UBE2I的异位表达可以促进乳腺癌细胞的侵袭和转移,包括骨转移 [39] 。UBE2C也是一个泛素相关基因,它编码破坏有丝分裂周期蛋白和细胞周期进展所需的蛋白质,据报道在乳腺癌的增殖和迁移中高度表达 [57] 。
Wnt信号通路是调控细胞增殖和分化的重要途径,大量研究表明Wnt信号通路与包括乳腺癌骨转移在内的骨恶性肿瘤密切相关 [65] [66] 。作为Wnt信号通路的一部分,CTNNB1已被证实在骨破坏过程中异常表达,这对乳腺癌骨转移的发生有重要意义 [11] 。CTNNB1编码的蛋白质是组成粘附连接(AJs)的复合蛋白的一部分,在骨肿瘤转移的骨病变阶段是必不可少的 [12] 。另一个参与Wnt信号通路的基因EGF,编码一种促进多种细胞类型生长、增殖和分化的蛋白质,EGF在患者血清中的表达对乳腺癌骨转移瘤的早期诊断具有重要价值 [30] [31] 。
在原癌基因家族中,HRAS是一种常见的致癌基因,已被报道为乳腺腺肌上皮瘤的驱动因素。体外实验证明,HRAS在乳腺癌骨转移过程中过度表达 [15] [16] 。作为乳腺癌的潜在治疗靶点,MYC编码一种核磷蛋白,在细胞周期进程、凋亡和细胞转化中发挥作用。MYC作为侵袭性和转移性乳腺癌的靶基因,其基因易位与骨微环境的修饰过程有关 [19] [20] 。另一个原癌基因MDM2参与了许多癌症的发病机制,包括乳腺癌的侵袭和转移过程。基质金属蛋白酶(MMP9)是一种增强肿瘤侵袭性的重要酶,MDM2促进了MMP9的高表达 [42] [43] 。MMP9是一种增加肿瘤侵袭性的重要酶。根据最近的研究,MDM2促进乳腺癌的侵袭和转移。对于另一个原癌基因JUN,研究表明该基因通过介导骨吸收过程中生长因子的释放增强了转移性乳腺癌细胞在骨骼中的生长和存活 [21] [22] 。
CDC37、CDC5L、CDK4、PLK1和CDC20是与细胞周期调控相关的基因。根据临床数据分析,PLK1和CDC20的表达与乳腺癌患者的无病生存率(DFS)密切相关。因此,PLK1和CDC20在促进乳腺肿瘤形成中发挥作用,并可能作为潜在的肿瘤诊断标志物和个体治疗靶标 [51] [52] 。CDK4是功能性丝氨酸/苏氨酸蛋白激酶家族的成员,参与细胞周期调节。根据其表达谱,CDK4在乳腺癌骨转移过程中被过度表达,作为乳腺癌骨转移的治疗靶点,其下调被认为是判断预后的合适指标 [17] [18] 。此外,CDC37通过稳定突变和过度表达的致癌激酶的受损结构来介导癌变和维持恶性表型,已被证实参与乳腺癌骨转移的功能途径 [13] [14] 。
对于其他一些重要的基因,CREBBP已被广泛报道与骨髓白血病的发展有关。在白血病的发病过程中,骨髓和骨组织的微环境发生了剧烈变化。作为骨组织的功能调节因子,CREBBP已被证实在乳腺癌骨转移过程中参与骨微环境的修饰过程,证实其在肿瘤转移过程中的重要作用 [40] [41] 。CD44上调了基质金属蛋白酶(MMPs)的表达,MMPs可以促进细胞从骨髓窦外渗,有利于乳腺癌细胞通过新生血管进入血液循环b并在乳腺癌骨转移中发挥特定作用 [27] [28] [29] 。BMP7编码TGF-β蛋白超家族的分泌配体,其诱导异位骨形成并促进骨转移。此外,BMP7参与了乳腺癌细胞的EMT过程,被报道直接参与乳腺癌脑转移的进程 [37] [38] 。鉴于NOTCH1在乳腺癌转移中的重要作用,它被认为是乳腺癌的核心治疗靶点。根据最近的文献研究,通过抑制NOTCH1信号通路,可以抑制乳腺癌细胞的骨转移 [48] [49] [50] 。
在尚未被验证的4个基因中,有3个存在于上述的基因家族中。其中,UBB和RPS27A是与泛素相关的基因,已发现它们在骨髓中广泛表达。通过降低UBB降低泛素水平,是一种潜在的癌症治疗方法 [67] 。RPS27A调节细胞周期进程并抑制白血病细胞的凋亡,暗示其在骨微环境的潜在作用 [68] [69] 。CDC5L是一个参与细胞周期调控的基因,最近的研究发现它是骨肉瘤和宫颈肿瘤的候选致病基因 [70] [71] 。另一个基因POLR2A是有关细胞存活的重要基因,已被确定为人类三阴性乳腺癌的精确靶标 [72] ,表明其在乳腺癌进展中的作用。
根据文献挖掘的结果及分析表明,通过该方法鉴定出的大多数潜在关键基因已被证实有助于乳腺癌骨转移的进展,这证明了所提出的综合方法可能是预测肿瘤转移相关基因和研究肿瘤转移机制的有用工具。
文章引用
田智萌,张子豪. 基于网络方法确定乳腺癌转移相关基因
Network-Based Method for the Identification of Genes Related to Cancer Metastasis[J]. 理论数学, 2023, 13(05): 1267-1280. https://doi.org/10.12677/PM.2023.135130
参考文献
- 1. Steeg, P.S. (2006) Tumor Metastasis: Mechanistic Insights and Clinical Challenges. Nature Medicine, 12, 895-904. https://doi.org/10.1038/nm1469
- 2. Chen, E.I. and Yates III, J.R. (2006) Maspin and Tumor Metastasis. IUBMB Life, 58, 25-29. https://doi.org/10.1080/15216540500531721
- 3. Jiang, M., Chen, Y., Zhang, Y., Chen, L., Zhang, N., Huang, T., Cai, Y.-D. and Kong, X. (2013) Identification of Hepatocellular Carcinoma Related Genes with k-th Shortest Paths in a Protein-Protein Interaction Network. Molecular BioSystems, 9, 2720-2728. https://doi.org/10.1039/c3mb70089e
- 4. Guo, W., Shang, D.-M., Cao, J.-H., Feng, K., He, Y.-C., Jiang, Y., Wang, S. and Gao, Y.-F. (2017) Identifying and Analyzing Novel Epilepsy-Related Genes Using Random Walk with Restart Algorithm. BioMed Research International, 2017, Article ID: 6132436. https://doi.org/10.1155/2017/6132436
- 5. Luo, H., Wang, J., Li, M., Luo, J., Ni, P., Zhao, K., Wu, F.-X. and Pan, Y. (2018) Computational Drug Repositioning with Random Walk on a Heterogeneous Network. IEEE/ACM Transactions on Computational Biology and Bioinformatics, 16, 1890-1900. https://doi.org/10.1109/TCBB.2018.2832078
- 6. Yu, W., Zhang, Z., Min, D., Yang, Q., Du, X., Tang, L., Lin, F., Sun, Y., Zhao, H. and Zheng, S. (2014) Mediator of RNA Polymerase II Transcription Subunit 19 Promotes Osteo-sarcoma Growth and Metastasis and Associates with Prognosis. European Journal of Cancer, 50, 1125-1136. https://doi.org/10.1016/j.ejca.2014.01.030
- 7. Christgen, M., Bartels, S., Luft, A., Persing, S., Henkel, D., Leh-mann, U. and Kreipe, H. (2018) Activating Human Epidermal Growth Factor Receptor 2 (HER2) Gene Mutation in Bone Metastases from Breast Cancer. Virchows Archiv, 473, 577-582. https://doi.org/10.1007/s00428-018-2414-1
- 8. Fu, W., Wu, H., Cheng, Z., Huang, S. and Rao, H. (2018) The Role of Katanin p60 in Breast Cancer Bone Metastasis. Oncology Letters, 15, 4963-4969. https://doi.org/10.3892/ol.2018.7942
- 9. Kim, J., Kim, Y., Choi, H., Kwon, A., Jekarl, D.W., Lee, S., Jang, W., Chae, H., Kim, J.R. and Kim, J.M. (2018) Ubiquitin C Decrement Plays a Pivotal Role in Replicative Senescence of Bone Marrow Mesenchymal Stromal Cells. Cell Death & Disease, 9, Article No. 139. https://doi.org/10.1038/s41419-017-0032-5
- 10. Najar, M., Fayyad-Kazan, H., Faour, W.H., Badran, B., Journe, F. and Lagneaux, L. (2017) Breast Cancer Cells and Bone Marrow Mesenchymal Stromal Cells: A Regulated Modulation of the Breast Tumor in the Context of Immune Response. Inflammation Research, 66, 129-139. https://doi.org/10.1007/s00011-016-1000-8
- 11. Johnson, R.W., Merkel, A.R., Page, J.M., Ruppender, N.S., Guelcher, S.A. and Sterling, J.A. (2014) Wnt Signaling Induces Gene Expression of Factors Associated with Bone De-struction in Lung and Breast Cancer. Clinical & Experimental Metastasis, 31, 945-959. https://doi.org/10.1007/s10585-014-9682-1
- 12. Tulotta, C. and Ottewell, P. (2018) The Role of IL-1B in Breast Cancer Bone Metastasis. Endocrine-Related Cancer, 25, R421-R434. https://doi.org/10.1530/ERC-17-0309
- 13. Hofer, E.L., Russa, V.L., Honegger, A.E., Bullorsky, E.O., Bordenave, R.H. and Chasseing, N.A. (2005) Alteration on the Expression of IL-1, PDGF, TGF-β, EGF, and FGF Receptors and c-Fos and c-Myc Proteins in Bone Marrow Mesenchymal Stroma Cells from Advanced Untreated Lung and Breast Cancer Patients. Stem Cells and Development, 14, 587-594. https://doi.org/10.1089/scd.2005.14.587
- 14. El Hamidieh, A., Grammatikakis, N. and Patsavoudi, E. (2012) Cell Surface Cdc37 Participates in Extracellular HSP90 Mediated Cancer Cell Invasion. PLOS ONE, 7, e42722. https://doi.org/10.1371/journal.pone.0042722
- 15. Holen, I., Nutter, F., Wilkinson, J., Evans, C., Avgoustou, P. and Ottewell, P.D. (2015) Human Breast Cancer Bone Metastasis in Vitro and in Vivo: A Novel 3D Model System for Studies of Tumour Cell-Bone Cell Interactions. Clinical & Ex-perimental Metastasis, 32, 689-702. https://doi.org/10.1007/s10585-015-9737-y
- 16. Nutter, F., Holen, I., Brown, H.K., Cross, S.S., Evans, C.A., Walker, M., Coleman, R.E., Westbrook, J.A., Selby, P.J. and Brown, J.E. (2014) Dif-ferent Molecular Profiles Are Associated with Breast Cancer Cell Homing Compared with Colonisation of Bone: Evi-dence Using a Novel Bone-Seeking Cell Line. Endocrine-Related Cancer, 21, 327-341. https://doi.org/10.1530/ERC-13-0158
- 17. Zhou, R., Frum, R., Deb, S. and Deb, S.P. (2005) The Growth Arrest Function of the Human Oncoprotein Mouse Double Minute-2 Is Disabled by Downstream Mutation in Cancer Cells. Cancer Research, 65, 1839-1848. https://doi.org/10.1158/0008-5472.CAN-03-3755
- 18. Martinez, R., Blasina, A., Hallin, J.F., Hu, W., Rymer, I., Fan, J., Hoffman, R.L., Murphy, S., Marx, M. and Yanochko, G. (2015) Mitotic Checkpoint Kinase Mps1 Has a Role in Normal Physiology Which Impacts Clinical Utility. PLOS ONE, 10, e0138616. https://doi.org/10.1371/journal.pone.0138616
- 19. Liu, H., Radisky, D.C., Yang, D., Xu, R., Radisky, E.S., Bissell, M.J. and Bishop, J.M. (2012) MYC Suppresses Cancer Metastasis by Direct Transcriptional Silencing of α v and β 3 Integrin Subunits. Nature Cell Biology, 14, 567-574. https://doi.org/10.1038/ncb2491
- 20. Zhao, J. and Jiang, G. (2018) MiR-4282 Inhibits Proliferation, Invasion and Metastasis of Human Breast Cancer by Targeting Myc. European Review for Medical and Pharmacological Sciences, 22, 8763-8771.
- 21. Nasrazadani, A. and Van den Berg, C. (2010) Abstract P4-06-02: Systemic Deletion of c-Jun N-Terminal Kinase 1 or 2 Is Protective in Breast Cancer Bone Metastasis. Cancer Research, 70, P4-06-02. https://doi.org/10.1158/0008-5472.SABCS10-P4-06-02
- 22. Qiao, H., Cui, Z., Yang, S., Ji, D., Wang, Y., Yang, Y., Han, X., Fan, Q., Qin, A. and Wang, T. (2017) Targeting Osteocytes to Attenuate Early Breast Cancer Bone Me-tastasis by Theranostic Upconversion Nanoparticles with Responsive Plumbagin Release. ACS Nano, 11, 7259-7273. https://doi.org/10.1021/acsnano.7b03197
- 23. Gayer, C.P., Craig, D.H., Flanigan, T.L., Reed, T.D., Cress, D.E. and Basson, M.D. (2010) ERK Regulates Strain-Induced Migration and Proliferation from Different Subcellular Locations. Journal of Cellular Biochemistry, 109, 711-725. https://doi.org/10.1002/jcb.22450
- 24. Zhao, J., Chen, H.-F. and Deng, H.-Y. (2009) Effects of Inhibiting JAK on Invasion and Metastasis of the Human Breast Cancer Cells through ERK Signaling Transduction Pathway. Chinese Journal of Cancer Research, 21, 32-36. https://doi.org/10.1007/s11670-009-0032-9
- 25. Li, Y.-B., Zhang, J.-P., Yao, W., Wang, H., Tang, J.-M. and Yang, J.-Y. (2010) VEGF Promote Migration of Bone Marrow-Derived Mesenchymal Stem Cells through PKC Signal Pathway. Journal of Yunyang Medical College, No. 3, 2.
- 26. Doucet, M., Kominsky, S., Nell, D. and Weber, K. (2005) TGF-Beta Stimulates IL-6, PDGF-AA, and VEGF in Renal Cell Carcinoma Bone Metastasis. Journal of Bone and Mineral Research, 20, S33.
- 27. Hill, A., McFarlane, S., Johnston, P. and Waugh, D. (2008) CD44 Signalling Increases Cathepsin K and MT1MMP Expression to Potentiate Breast Cancer Cell Invasion through Collagen I. Breast Cancer Research, 10, P33. https://doi.org/10.1186/bcr1917
- 28. Hill, A., McFarlane, S., Johnston, P. and Waugh, D. (2006) CD44 Regulates the Expression of the Cysteine Protease Cathepsin K: Implications for Bone Metastasis of Breast Cancer. European Journal of Cancer Supplements, 4, 17. https://doi.org/10.1016/S1359-6349(06)70049-0
- 29. Liu, Y., Qing, H., Su, X., Wang, C., Li, Z. and Liu, S. (2015) Association of CD44 Gene Polymorphism with Survival of NSCLC and Risk of Bone Metastasis. Medical Science Monitor: International Medical Journal of Experimental and Clinical Research, 21, 2694-2700. https://doi.org/10.12659/MSM.894357
- 30. Chang, Y.-S., Chen, W.-Y., Yin, J. J., Sheppard-Tillman, H., Huang, J. and Liu, Y.-N. (2015) EGF Receptor Promotes Prostate Cancer Bone Metastasis by Downregulating miR-1 and Acti-vating TWIST1. Cancer Research, 75, 3077-3086. https://doi.org/10.1158/0008-5472.CAN-14-3380
- 31. Wang, S.-J., Saadi, W., Lin, F., Nguyen, C.M.-C. and Jeon, N.L. (2004) Differential Effects of EGF Gradient Profiles on MDA-MB-231 Breast Cancer Cell Chemotaxis. Experi-mental Cell Research, 300, 180-189. https://doi.org/10.1016/j.yexcr.2004.06.030
- 32. Nasrazadani, A. and Van Den Berg, C.L. (2011) c-Jun N-Terminal Kinase 2 Regulates Multiple Receptor Tyrosine Kinase Pathways in Mouse Mammary Tumor Growth and Metastasis. Genes & Cancer, 2, 31-45. https://doi.org/10.1177/1947601911400901
- 33. Xu, J., Ying, Y., Xiong, G., Lai, L., Wang, Q. and Yang, Y. (2019) Amyloid β Precursor Protein Silencing Attenuates Epithelial-Mesenchymal Transition of Nasopharyngeal Car-cinoma Cells via Inhibition of the MAPK Pathway. Molecular Medicine Reports, 20, 409-416. https://doi.org/10.3892/mmr.2019.10293
- 34. Lim, S., Yoo, B.K., Kim, H.-S., Gilmore, H.L., Lee, Y., Lee, H.-P., Kim, S.-J., Letterio, J. and Lee, H.-G. (2014) Amyloid-β Precursor Protein Promotes Cell Proliferation and Motility of Advanced Breast Cancer. BMC Cancer, 14, Article No. 928. https://doi.org/10.1186/1471-2407-14-928
- 35. Sohara, Y., Peng, H., Minkin, C., Nolta, J., Wang, H., Dorey, F., Reynolds, P., Shimada, H. and DeClerck, Y. (2004) Contribution of Bone Marrow Mesenchymal Stem Cells and IL-6 to Bone Metastasis. Journal of Bone and Mineral Research, 19, 1577.
- 36. Wu, C., Chen, M., Sun, Z., Ye, Y., Han, X., Qin, Y. and Liu, S. (2019) Wenshen Zhuanggu Formula Mitigates Breast Cancer Bone Metastasis through the Signaling Crosstalk among the Jagged1/Notch, TGF-β and IL-6 Signaling Pathways. Journal of Ethnopharmacology, 232, 145-154. https://doi.org/10.1016/j.jep.2018.12.023
- 37. Kijewska, M., Viski, C., Turrell, F., Fitzpatrick, A., van Weverwijk, A., Gao, Q., Iravani, M. and Isacke, C.M. (2019) Using an In-Vivo Syngeneic Spontaneous Metastasis Model Identifies ID2 as a Promoter of Breast Cancer Colonisation in the Brain. Breast Cancer Research, 21, Article No. 4. https://doi.org/10.1186/s13058-018-1093-9
- 38. Buijs, J.T., Henriquez, N.V., van Overveld, P.G., van der Horst, G., Ten Dijke, P. and van der Pluijm, G. (2007) TGF-β and BMP7 Interactions in Tumour Progression and Bone Metastasis. Clinical & Experimental Metastasis, 24, 609-617. https://doi.org/10.1007/s10585-007-9118-2
- 39. Zhu, S., Sachdeva, M., Wu, F., Lu, Z. and Mo, Y.-Y. (2010) Ubc9 Promotes Breast Cell Invasion and Metastasis in a Sumoylation-Independent Manner. Oncogene, 29, 1763-1772. https://doi.org/10.1038/onc.2009.459
- 40. Teramachi, J., Silbermann, R., Yang, P., Zhao, W., Mohammad, K.S., Guo, J. anderson, J.L., Zhou, D., Feng, R., Myint, K.Z., Maertz, N., Beumer, J.H., Eiseman, J.L., Windle, J.J., Xie, X.Q., Roodman, G.D. and Kurihara, N. (2016) Blocking the ZZ Domain of Sequestosome1/p62 Suppresses Myeloma Growth and Osteoclast Formation in Vitro and Induces Dramatic Bone Formation in Myeloma-Bearing Bones in Vivo. Leukemia, 30, 390-398. https://doi.org/10.1038/leu.2015.229
- 41. Fan, Q.M., Yue, B., Bian, Z.Y., Xu, W.T., Tu, B., Dai, K.R., Li, G. and Tang, T.T. (2012) The CREB-Smad6-Runx2 Axis Contributes to the Impaired Osteogenesis Potential of Bone Marrow Stromal Cells in Fibrous Dysplasia of Bone. The Journal of Pathology, 228, 45-55. https://doi.org/10.1002/path.4033
- 42. Chen, X., Qiu, J., Yang, D., Lu, J., Yan, C., Zha, X. and Yin, Y. (2013) MDM2 Promotes Invasion and Metastasis in Invasive Ductal Breast Carcinoma by Inducing Matrix Metalloproteinase-9. PLOS ONE, 8, e78794. https://doi.org/10.1371/journal.pone.0078794
- 43. Qian, G., Hao, S., Yang, D. and Meng, Q. (2015) P15, MDM2, NF-κB, and Bcl-2 Expression in Primary Bone Tumor and Correlation with Tumor Formation and Metastasis. International Journal of Clinical and Experimental Pathology, 8, 14885-14892.
- 44. Cizkova, M., Vacher, S., Meseure, D., Trassard, M., Susini, A., Mlcuchova, D., Callens, C., Rouleau, E., Spyratos, F. and Lidereau, R. (2013) PIK3R1 Underexpression Is an Independent Prognostic Marker in Breast Cancer. BMC Cancer, 13, Article No. 545. https://doi.org/10.1186/1471-2407-13-545
- 45. Van Allen, E., Foye, A., Wagle, N., Kim, W., Carter, S., McKenna, A., Simko, J., Garraway, L. and Febbo, P. (2014) Successful Whole-Exome Sequencing from a Prostate Cancer Bone Metastasis Biopsy. Prostate Cancer and Prostatic Diseases, 17, 23-27. https://doi.org/10.1038/pcan.2013.37
- 46. Ruiz-Garcia, E., Scott, V., Machavoine, C., Bidart, J., Lacroix, L., Delaloge, S. and Andre, F. (2010) Gene Expression Profiling Identifies Fibronectin 1 and CXCL9 as Candidate Biomarkers for Breast Cancer Screening. British Journal of Cancer, 102, 462-468. https://doi.org/10.1038/sj.bjc.6605511
- 47. von Au, A., Vasel, M., Tiwari, S., Glüer, C. and Nakchbandi, I. (2010) Source and Significance of Circulating Fibronectin in Breast Cancer-Induced Bone Metastasis. Bone, 47, S113. https://doi.org/10.1016/j.bone.2010.04.240
- 48. Wang, J., Fu, L., Gu, F. and Ma, Y. (2011) Notch1 Is Involved in Migration and Invasion of Human Breast Cancer Cells. Oncology Reports, 26, 1295-1303.
- 49. Pei, J. and Wang, B. (2015) Notch-1 Promotes Breast Cancer Cells Proliferation by Regulating LncRNA GAS5. International Journal of Clinical and Experimental Medicine, 8, 14464-14471.
- 50. Kong, P., Chen, L., Yu, M., Tao, J., Liu, J., Wang, Y., Pan, H., Zhou, W. and Wang, S. (2018) miR-3178 Inhibits Cell Proliferation and Metastasis by Targeting Notch1 in Triple-Negative Breast Cancer. Cell Death & Disease, 9, Article No. 1059. https://doi.org/10.1038/s41419-018-1091-y
- 51. Wang, L., Zhang, J., Wan, L., Zhou, X., Wang, Z. and Wei, W. (2015) Targeting Cdc20 as a Novel Cancer Therapeutic Strategy. Pharmacology & Therapeutics, 151, 141-151. https://doi.org/10.1016/j.pharmthera.2015.04.002
- 52. Liu, G., Chen, Y., Qi, F., Jia, L., Lu, X.A., He, T., Fu, Y., Li, L. and Luo, Y. (2015) Specific Chemotherapeutic Agents Induce Metastatic Behaviour through Stromal- and Tumour-Derived Cytokine and Angiogenic Factor Signalling. The Journal of Pathology, 237, 190-202. https://doi.org/10.1002/path.4564
- 53. Li, R., Peng, C., Zhang, X., Wu, Y., Pan, S. and Xiao, Y. (2017) Roles of Arf6 in Cancer Cell Invasion, Metastasis and Proliferation. Life Sciences, 182, 80-84. https://doi.org/10.1016/j.lfs.2017.06.008
- 54. Xu, R., Zhang, Y., Gu, L., Zheng, J., Cui, J., Dong, J. and Du, J. (2015) Arf6 Regulates EGF-Induced Internalization of E-Cadherin in Breast Cancer Cells. Cancer Cell International, 15, Article No. 11. https://doi.org/10.1186/s12935-015-0159-3
- 55. Morrison, M.M., Young, C.D., Wang, S., Sobolik, T., Sanchez, V.M., Hicks, D.J., Cook, R.S. and Brantley-Sieders, D.M. (2015) mTOR Directs Breast Morphogenesis through the PKC-alpha-Rac1 Signaling Axis. PLoS Genetics, 11, e1005291. https://doi.org/10.1371/journal.pgen.1005291
- 56. Zheng, H. and Kang, Y. (2015) Cradle of Evil: Osteogenic Niche for Early Bone Metastasis. Cancer Cell, 27, 153-155. https://doi.org/10.1016/j.ccell.2015.01.006
- 57. Chou, C.-P., Huang, N.-C., Jhuang, S.-J., Pan, H.-B., Peng, N.-J., Cheng, J.-T., Chen, C.-F., Chen, J.-J. and Chang, T.-H. (2014) Ubiquitin-Conjugating Enzyme UBE2C Is Highly Expressed in Breast Microcalcification Lesions. PLOS ONE, 9, e93934. https://doi.org/10.1371/journal.pone.0093934
- 58. Carthew, R.W., Chodosh, L.A. and Sharp, P.A. (1985) An RNA Polymerase II Transcription Factor Binds to an Upstream Element in the Adenovirus Major Late Promoter. Cell, 43, 439-448. https://doi.org/10.1016/0092-8674(85)90174-6
- 59. Hu, K.-F., Kong, X.-Y., Zhong, M.-C., Wan, H.-Y., Lin, N. and Pei, X.-H. (2017) Brucine Inhibits Bone Metastasis of Breast Cancer Cells by Suppressing Jagged1/Notch1 Signaling Pathways. Chinese Journal of Integrative Medicine, 23, 110-116. https://doi.org/10.1007/s11655-016-2647-2
- 60. Chen, X., Pei, Z., Peng, H. and Zheng, Z. (2018) Exploring the Molecular Mechanism Associated with Breast Cancer Bone Metastasis Using Bioinformatic Analysis and Microarray Genetic Interaction Network. Medicine, 97, e12032. https://doi.org/10.1097/MD.0000000000012032
- 61. Wu, X., Sun, L., Wang, X., Su, P., Li, Z., Zhang, C., Wang, Y., Gao, P. and Ma, R. (2016) Breast Cancer Invasion and Metastasis by mPRα through the PI3K/Akt Signaling Pathway. Pathology & Oncology Research, 22, 471-476. https://doi.org/10.1007/s12253-015-0023-8
- 62. Kalantzaki, K., Koumakis, L., Bei, E.S., Zervakis, M., Potamias, G. and Kafetzopoulos, D. (2013) Experimental Model Construction and Validation of the ErbB Signaling Pathway. 13th IEEE International Conference on BioInformatics and BioEngineering, Chania, 10-13 November 2013, 1-4. https://doi.org/10.1109/BIBE.2013.6701557
- 63. Li, X.-Q., Du, X., Li, D.-M., Kong, P.-Z., Sun, Y., Liu, P.-F., Wang, Q.-S. and Feng, Y.-M. (2015) ITGBL1 Is a Runx2 Transcriptional Target and Promotes Breast Cancer Bone Metastasis by Activating the TGFβ Signaling Pathway. Cancer Research, 75, 3302-3313. https://doi.org/10.1158/0008-5472.CAN-15-0240
- 64. Drabsch, Y. and Ten Dijke, P. (2011) TGF-β Signaling in Breast Cancer Cell Invasion and Bone Metastasis. Journal of Mammary Gland Biology and Neoplasia, 16, 97-108. https://doi.org/10.1007/s10911-011-9217-1
- 65. Ferreira Tojais, N., Peghaire, C., Franzl, N., Larrieu-Lahargue, F., Jaspard, B., Reynaud, A., Moreau, C., Couffinhal, T., Duplàa, C. and Dufourcq, P. (2014) Frizzled7 Controls Vascular Permeability through the Wnt-Canonical Pathway and Cross-Talk with Endothelial Cell Junction Complexes. Cardiovascular Research, 103, 291-303. https://doi.org/10.1093/cvr/cvu133
- 66. Chen, Y., Shi, H.Y., Stock, S.R., Stern, P.H. and Zhang, M. (2011) Regulation of Breast Cancer-Induced Bone Lesions by β-Catenin Protein Signaling. Journal of Biological Chemistry, 286, 42575-42584. https://doi.org/10.1074/jbc.M111.294595
- 67. Oh, C., Park, S., Lee, E.K. and Yoo, Y.J. (2013) Downregulation of Ubiquitin Level via Knockdown of Polyubiquitin Gene Ubb as Potential Cancer Therapeutic Intervention. Scientific Reports, 3, Article No. 2623. https://doi.org/10.1038/srep02623
- 68. He, R., Yang, N., Zhang, P., Liu, J., Li, J., Zhou, F. and Zhang, W. (2017) Identification and Expression of MMSA-8, and Its Clinical Significance in Multiple Myeloma. Oncology Reports, 37, 3235-3243. https://doi.org/10.3892/or.2017.5609
- 69. Wang, H., Yu, J., Zhang, L., Xiong, Y., Chen, S., Xing, H., Tian, Z., Tang, K., Wei, H. and Rao, Q. (2014) RPS27a Promotes Proliferation, Regulates Cell Cycle Progression and Inhibits Apoptosis of Leukemia Cells. Biochemical and Biophysical Research Communications, 446, 1204-1210. https://doi.org/10.1016/j.bbrc.2014.03.086
- 70. Chen, W., Zhang, L., Wang, Y., Sun, J., Wang, D., Fan, S., Ban, N., Zhu, J., Ji, B. and Wang, Y. (2016) Expression of CDC5L Is Associated with Tumor Progression in Gliomas. Tumor Biology, 37, 4093-4103. https://doi.org/10.1007/s13277-015-4088-5
- 71. Lu, X.-Y., Lu, Y., Zhao, Y.-J., Jaeweon, K., Kang, J., Li, X.-N., Ge, G., Meyer, R., Perlaky, L. and Hicks, J. (2008) Cell Cycle Regulator Gene CDC5L, a Potential Target for 6p12-p21 Amplicon in Osteosarcoma. Molecular Cancer Research, 6, 937-946. https://doi.org/10.1158/1541-7786.MCR-07-2115
- 72. Xu, J., Liu, Y., Li, Y., Wang, H., Stewart, S., Van der Jeught, K., Agarwal, P., Zhang, Y., Liu, S. and Zhao, G. (2019) Precise Targeting of POLR2A as a Therapeutic Strategy for Human Triple Negative Breast Cancer. Nature Nanotechnology, 14, 388-397. https://doi.org/10.1038/s41565-019-0381-6
