Advances in Microbiology
Vol.3 No.03(2014), Article ID:14257,8 pages
DOI:10.12677/AMB.2014.33009
Pyrosequencing Analysis Reveals Abundance and Community Composition of Denitrifying Bacteria in Xilin River Sludge
1College of Environment and Resources, Inner Mongolia University, Hohhot
2College of Life Sciences, Inner Mongolia University, Hohhot
3Sino-US Center for Conservation, Energy Science in Inner Mongolia, Hohhot
Email: hot-yjl@163.com, *ndzj@imu.edu.cn
Copyright © 2014 by authors and Hans Publishers Inc.
This work is licensed under the Creative Commons Attribution International License (CC BY).
http://creativecommons.org/licenses/by/4.0/



Received: Aug. 10th, 2014; revised: Sep. 12th, 2014; accepted: Sep. 20th, 2014
ABSTRACT
Nitrogen loss caused by denitrifying bacteria is an important link in the global carbon-nitrogen cycles, whose community compositions and abundance may have a role in regulating emissions and forms of gaseous nitrogen. Based on 454 pyrosequencing of the 16S rRNA gene, as well as the reported denitrifying bacterial genera mentioned in many literatures, we detected community compositions and abundance of denitrifying bacterial communities in Xilin River sludge. The results showed that 5744 quality reads were grouped into 2263 OTUs at genus level (0.05 distance). After alignment with known sequences deposited in GenBank, the unclassified bacteria genera accounted for 57.49%. 204 classified bacterial genera covered 20 denitrifying bacteria genera, including 1.40% of Flavobacterium, 1.23% of Hydrogenophaga, 0.19% of Bacillus and Thauera, 0.17% of Thiobacillus, 0.15% of Hyphomicrobium, etc. Among them, Flavobacterium and Hydrogenophaga were the most dominant denitrifying bacterial genera in Xilin River bottom sludge, accounting for 69.03% of the total denitrifying bacterial genera. Correlation analysis showed that Flavobacterium preferred to attach to coarse sand grains in relatively high pH conditions, promoting the emission of CH4 and inhibiting the production of N2O and CO2. In opposition, Hydrogenophaga showed special preference to fine silt grains in rich nutrition conditions, promoting the emission of N2O and CO2 and inhibiting the production of CH4.
Keywords:Pyrosequencing Analysis, Denitrifying Bacteria, Community Compositions, Abundance, Xilin River Sludge

高通量技术解析锡林河底泥反硝化菌群
组成及丰度
于景丽1,2,3,范雅慧1,高晓霞1,史昊先2,赵 吉1,3*
1内蒙古大学环境与资源学院,呼和浩特
2内蒙古大学生命科学学院,呼和浩特
3中美生态、能源及可持续性科学内蒙古研究中心,呼和浩特
Email: hot-yjl@163.com, *ndzj@imu.edu.cn
收稿日期:2014年8月10日;修回日期:2014年9月12日;录用日期:2014年9月20日

摘 要
反硝化菌驱动的脱氮作用是全球氮循环的重要环节,其群落组成及丰度对气态氮产生的类型和排放量有调控作用。本文结合文献报道的反硝化细菌属,基于16S rRNA基因454焦磷酸测序检测了锡林河底泥中反硝化细菌群落结构组成和丰度。结果发现:5744条优化序列在“属”水平上聚类成2263个OTUs,与GenBank中已知序列比对后获得未分类的细菌属占57.49%。已分类的204个细菌属中20个为反硝化细菌属,包括Flavobacterium 1.40%,Hydrogenophaga 1.23%,Bacillus和Thauera 0.19%,Thiobacillus 0.17%,Hyphomicrobium 0.15%等。其中,黄杆菌属Flavobacterium和噬氢菌属Hydrogenophaga是锡林河底泥中最丰富的反硝化细菌,共占反硝化菌群总组成的69.03%。相关性分析表明:Flavobacterium更喜欢pH相对高的沙粒,促进CH4排放并抑制N2O和CO2产生;而Hydrogenophaga更喜欢营养丰富的细粒,促进N2O和CO2排放并抑制CH4产生。
关键词
焦磷酸测序,反硝化细菌,群落组成,丰度,锡林河底泥

1. 引言
以454焦磷酸Pyrosequencing和Illumina Solexa等为代表的高通量测序技术不但能定性研究微生物的群落组成及多样性,还能定量分析优势菌、常见菌和罕见菌的相对丰度,兼备微生物群落结构分析技术(DGGE和T-RFLP)[1] -[5] 和定量分析技术(qPCR)[1] [6] 两种功能,在研究复杂环境样品微生物群落结构组成、丰度及多样性研究中发挥了重要作用[7] -[10] 。
土壤及水体中N2O的排放主要是反硝化菌驱动的生物脱氮作用[6] [11] -[15] 。目前国内外关于反硝化菌的研究主要集中在菌株的分离培养、反硝化活力检测及氮素富集的污泥和污水的脱氮性能等方面[16] -[19] 。菌株种属的鉴定主要通过形态观察、生理生化特性及16S rRNA基因同源性分析来进行[16] -[19] 。迄今已发现60多个属的细菌具有反硝化功能,包括传统意义上的厌氧反硝化菌和目前备受关注的好氧反硝化菌[12] [13] [20] 。同时在土壤反硝化菌群丰度与N2O释放量间关系方面也取得重要进展[6] [14] [15] 。但天然河流底泥反硝化菌群组成、丰度及空间分布特征还鲜为人知。大量研究表明,反硝化菌群组成及丰缺直接影响土壤氮素的迁移和转化,并调控气态氮产生类型(N2O/N2)和排放量[11] [13] 。
环境样品中可培养的微生物不足总数的0.1%~10%。高通量测序为功能菌群的大量筛选提供了契机。本文基于传统16S rRNA基因序列分析建立的反硝化菌基因资源库,通过细菌16S rRNA基因454焦磷酸测序技术获得的大量数据,从“属”水平上挖掘具有反硝化功能的细菌群结构组成及丰度,为预测锡林河底泥氮素逸失和温室气体释放提供科学依据。
2. 材料与方法
2.1. 研究区概况及常规指标检测
锡林河是内蒙古高原半干旱区最具代表性的内陆河。样点设置在中国科学院内蒙古草原生态系统定位站西侧(43˚36'N~44˚29'N,115˚32'E~117˚12'E)。该区属温带半干旱大陆性季风气候,年均温−0.4℃,年均降水量336.9 mm。水生植被主要有水甜茅(Glyceria triflora)、散穗早熟禾(Poa subfastigiata)、灰脉苔草(Carex appendiculata)等。沿着河流走向多点采集河边腥臭底泥。用激光法、105℃烘干法、电位法、电导率法分别检测底泥的颗粒组成、水分含量(WC)、pH、水溶性盐分总量(DS)。用总有机碳分析仪、半微量凯氏蒸馏法、钼锑抗分光光度法分别测定总有机碳(TOC)、全氮(TN)、全磷(TP)。河边底泥中沙粒含量15% ± 1%,粉粒含量55% ± 2%左右,粘粒含量30% ± 1%。WC: 22.72% ± 1.83%, pH: 8.31 ± 0.15, DS: 0.71 ± 0.16 g∙kg−1, TOC: 6.45 ± 1.14 g∙kg−1, TN: 1.27 ± 0.34 g∙kg−1, TP: 0.33 ± 0.09 g∙kg−1。用Agilent 7890A气相色谱法检测N2O、CH4、CO2的浓度。待测气体浓度:Cs(ppm) = As*C0/A0,其中As:待测样品峰面积;C0:标气浓度(1.0 ppm);A0:标气峰面积。河边底泥N2O、CH4、CO2排放量依次为0.372 ± 0.014、2.227 ± 0.013、623.6 ± 3.9 ppm。
2.2. 河流底泥DNA的提取、PCR和焦磷酸测序
样品充分混匀后称取0.5 g按文献[21] 进行微生物基因组总DNA提取。利用16S rRNA基因V1-V3区引物进行PCR扩增。引物8F(5’-AGAGTTTGATCCTGGCTCAG-3’)加454-A接头(5’-cgtatcgcctccctcgcgccatcag-3’),533R(5’-TTACCGCGGCTGCTGGCAC-3’)加454-B接头引物序列(5’-ctatgcgccttgccagcccgctcag -3’)[10] 。每个样品加10 bp标签(barcode)序列以示区别。PCR反应体系(20 μL):DNA模板1 μL,FastPfu DNA聚合酶0.4 μL,5 × FastPfu缓冲液4 μL,2.5 m mol∙L−1 dNTPs 2 μL,正、反向引物(5 μ mol∙L−1)各0.4 μL,最后补超纯水到20 μL。PCR 反应条件:95℃预变性2 min;95℃变性30 s,55℃退火30 s,72℃延伸30 s,25个循环;72℃后延伸5 min。所有样品PCR产物经纯化和定量后送往上海美吉生物医药科技有限公司的罗氏基因组GS-FLX 454 Titanium测序平台(Roche, Mannheim, Germany)进行大规模平行测序。
2.3. 测序数据分析
将测序接头、标签和前引物序列去除后得到有效序列,进一步修剪、去除非特异性扩增片段、模糊碱基、单碱基高重复区以及长度过短的序列,得到长度>150 bp的优化序列。然后与SILVA数据库(http://www.arb-silva.de)中16S rRNA基因序列进行比对,去除嵌合体(Chimeric)序列,通过Mothur软件(http://www.mothur.org)进行距离的计算和 OTUs(operational taxonomic units,可操作分类单元)的聚类。登陆EMBL/GenBank/DDBJ核酸序列数据库(Accession Number: DRA000705)[10] ,基于OTUs在0.03、0.05、0.10水平上的遗传距离分别获得种、属、科水平上的细菌组成及单个菌的相对丰度[22] ,用Mothur软件Plot-rarefaction程序生成稀疏曲线图。
2.4. 数据的统计分析
采用Excel2007和Origin Lab 8.5软件(http://www.originlab.com)进行基本图形绘制。用SPSS16.0(SPSS Inc, Chicago, USA)软件进行Pearson相关性分析。表征群落中反硝化菌属丰富度、优势度和均匀度指数计算方法:1) Shannon-Wiener多样性指数(H′) H′= −∑PilnPi,2) +Simpson优势度指数(D) D = 1 − ∑(Pi)2,3) Pielou均匀度指数(Jp)Jp = −(∑PilnPi)/lnS,式中:S——所在群落的反硝化菌属数目;N——所在群落的所有反硝化菌属的个体数之和;Ni——所在群落内反硝化菌属i的个体数;Pi = Ni/N。
3. 结果与分析
3.1. 焦磷酸测序结果分析
基于16S rRNA基因焦磷酸测序分析,本研究先后获得6353条有效序列和5744条优化序列(Quality Reads)。图1为OTUs在0.03、0.05、0.10遗传差异的稀疏曲线图。图1稀疏曲线和表1中的覆盖度(Coverage)分别反映了样品测序的深度和拼接后序列在基因组中的覆盖度程度。选择0.05遗传距离生成的2263个OTUs经GenBank序列比对后,发现No_Rank菌群丰度高达总细菌组成的57.49%,说明在“属”水平上拼接后的序列在基因组中覆盖度不高,暗示很多未分类细菌的功能尚未揭示。已分类的204个细菌属累积丰度占总细菌组成的42.51%,结合文献大量报道的反硝化细菌属,本研究从已分类的204个菌属中共筛选出20个反硝化细菌属,占总细菌组成的3.81%(如图2)。
3.2. 反硝化细菌群落结构组成及丰度
20个反硝化菌属相对丰度分布范围为0.02%~1.40%,累积丰度占204个细菌属中的9.01%。依据相对丰度由高到低的顺序20个反硝化菌属群落组成排序如下:Flavobacterium 1.40%,Hydrogenophaga 1.23%,Bacillus和Thauera 0.19%,Thiobacillus 0.17%,Hyphomicrobium 0.15%,Stenotrophomonas 0.08%,Microbacterium和Alcaligenes 0.06%,Pseudomonas、Rhizobium和Herbaspirillum 0.04%,Sphingobacterium、Iamia、Acinetobacter、Rhodospirillaceae、Sorangium、Rhodococcus、Derxia和Zoogloea均为0.02%(图3)。

Figure 1. Rarefaction curve
图1. 稀疏曲线
表1. 样品的测序信息及多样性指标
DNB: denitrifying bacteria.

Figure 2. Proportion of different taxonomic groups
图2. 不同类群的分布比例
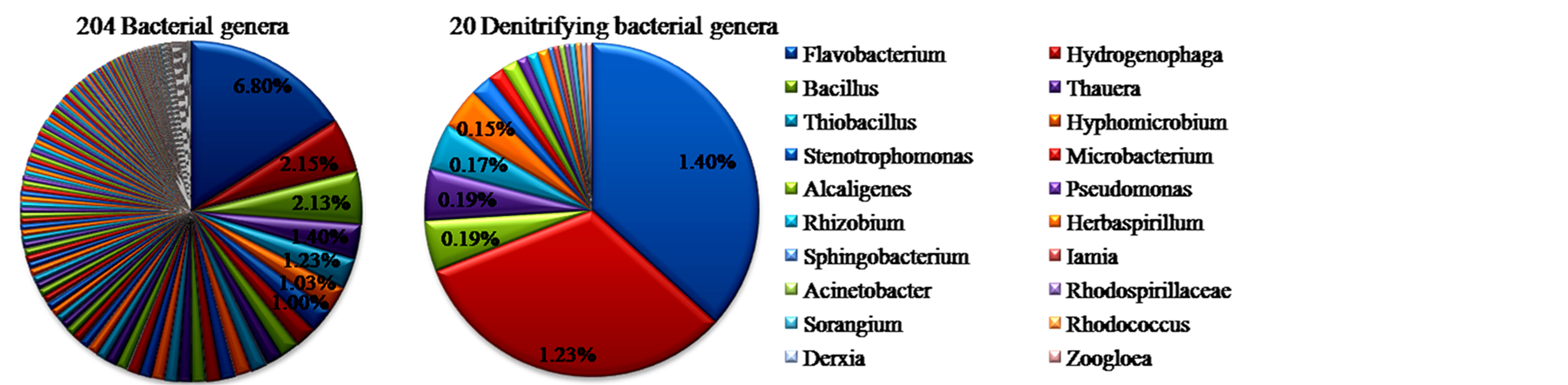
Note◎ 20 denitrifying bacterial genera were as followed:1. Flavobacterium黄杆菌属,2. Hydrogenophaga噬氢菌属,3. Bacillus芽孢杆菌属,4. Thauera索氏菌属,5. Thiobacillus硫杆状菌属,6. Hyphomicrobium生丝微菌属,7. Stenotrophomonas嗜麦芽寡养单胞菌属,8. Microbacterium微杆菌属,9. Alcaligenes产碱菌属,10. Pseudomonas假单胞菌属,11. Rhizobium根瘤菌属,12. Herbaspirillum草螺菌属,13. Sphingobacterium鞘氨醇杆菌属,14. Iamia,15. Acinetobacter不动杆菌属,16. Rhodospirillaceae_光合红螺菌科,17. Sorangium粘细菌属,18. Rhodococcus红球菌属,19. Derxia德克斯氏菌属,20. Zoogloea动胶杆菌属.
Note○ 204 bacterial genera were as followed:

Figure 3. Relative abundance and community compositions of bacteria and denitrifying bacteria in Xilin River Sludge
图3. 锡林河底泥中细菌和反硝化细菌群落组成及相对丰度
3.3. 优势反硝化细菌属及相应高级分类单元的组成及丰度
本研究依据总细菌属中单个菌相对丰度的高低,将反硝化细菌划分为3个等级:低(≤0.02%),中等(0.02%~0.50%),高(0.50%~1.00%),极高(≥1.00%)。基于反硝化细菌丰度分配等级,确定丰度高(0.50%~1.00%)和极高(≥1.00%)的反硝化菌属作为优势菌。河流底泥中只发现2个优势反硝化细菌,分别是黄杆菌属Flavobacterium和噬氢菌属Hydrogenophaga,两者分别占反硝化细菌群总组成的36.75%和32.28%(图4),合计占69.03%,接近反硝化菌群总组成的70%。考虑到两者的特殊性,我们逐级放大分类单元分析了门、纲、目、科的相对丰度,发现噬氢菌属Hydrogenophaga所对应的变形菌门Proteobacteria、变形菌纲β-proteobacteria、伯克氏菌目Burkholderiales、丛毛单胞菌科Comamonadaceae的相对丰度依次为44.21%、21.82%、12.41%、10.82%,Hydrogenophaga所在的门、纲、目和科都是细菌群落中最优势的组成部分。相比而言,黄杆菌属Flavobacterium所对应的拟杆菌门Bacteroidetes、黄杆菌纲Flavobacteria、黄杆菌目Flavobacteriales、黄杆菌科Flavobacteriaceae的相对丰度依次为15.77%、2.03%、2.03%、1.67%,也是细菌群落中较优势的组成部分。
4. 讨论
4.1. 高通量测序用于大量筛选反硝化等功能菌的优越性
通过16S rRNA基因焦磷酸测序,本研究一次性筛选出20个反硝化细菌属,真实体现了焦磷酸测序的高通量性及无需培养和克隆测序等繁琐步骤的快捷性。更为重要的是,高通量测序具有传统分子生物学的双重功能,可同步分析微生物群落结构组成和丰度[7] -[9] ,为筛选优势菌提供强有力的技术保障。
4.2. 反硝化优势菌的空间异质性成因及其群落构建机制
通过16S rRNA基因焦磷酸测序分析,获得反硝化菌的相对丰度最低0.02%,最高1.40%,确定相对丰度>1.0%的Flavobacterium和Hydrogenophaga是锡林河河边腥臭底泥中的优势反硝化菌。Park等[23] 通过DGGE技术发现生物膜反应器中的优势菌属Hydrogenophaga是不可培养的新型反硝化细菌属,能利用氢作为电子供体来移除硝酸盐。Calderer等[1] 发现碳氮比是决定细菌反硝化潜势的主要因子,当添加葡萄糖时Hydrogenophaga变成优势菌。Michaud等[24] 通过依赖和不依赖于PCR等多相分子技术研究南极洲浅水湖泊微生物的群落组成,也发现和本研究同样的优势菌Flavobacterium,说明这些优势菌不仅具
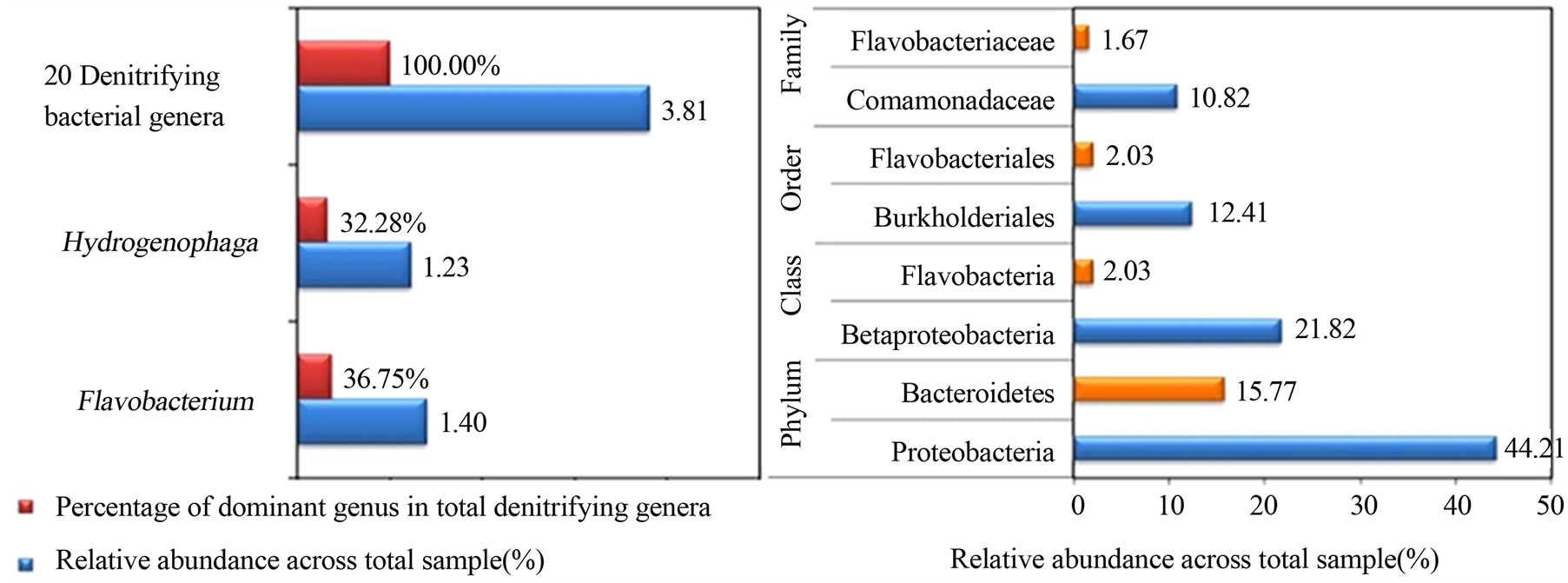
Figure 4. Dominant denitrifying bacterial genera and their corresponding Phyla, Classes, Orders, Families in Xilin River sludge
图4. 锡林河底泥中优势的细菌属和相应的门,纲,目,科
Table 2. Correlation analysis between environmental factors, greenhouse gas and dominant denitrifying bacterial genera in slurry and sandy sediments of Xilin River
表2. 河流泥、沙质沉积物中优势反硝化菌属与环境因子及温室气体的相关性分析
*Correlation is significant at the 0.05 level (2-tailed);*0.05水平上显著相关(两尾)。**Correlation is significant at the 0.01 level (2-tailed);**0.01水平上显著相关(两尾)。
有南北极淡水湖泊、池塘、河流、冰川的两极分布特征,还具有内陆河分布特征。这刷新了人们对优势反硝化菌主要存在于污水和活性污泥等氮污染环境的传统认识[3] [4] [16] -[19] [25] -[28] 。总体来讲,关于Hydrogenophaga和Flavobacterium作为反硝化优势细菌属的报道很少,“属”水平上的空间分布、生境特征及影响因素等尚无定论。但是Hydrogenophaga和Flavobacterium所对应的变形菌门Proteobacteria和拟杆菌门Bacteroidetes是门水平细菌群落中排名第一和第二的优势菌,两者相对丰度分别为44.21%和15.77%,累积丰度59.98%,接近细菌群落总组成的60%。这间接验证了Hydrogenophaga和Flavobacterium作为优势反硝化细菌属出现在河流底泥中的客观真实性。
本课题组通过前期对比研究发现锡林河中心沉积物以沙粒为主(沙粒含量 > 95%),自然含水量WC (17.95 ± 1%)、水溶性盐DS (0.30 ± 0.02 g∙kg−1)、总有机碳TOC (4.51 ± 0.14 g∙kg−1)、全氮TN (0.44 ± 0.05 g∙kg−1)、全磷TN (0.03 ± 0.01 g∙kg−1)显著低于河边底泥(P < 0.01),但pH略高于河边底泥。温室气体对比研究发现,河边泥质沉积物中N2O和CH4排放量与河中心沙质沉积物N2O (0.363 ± 0.007 ppm)和CH4 (2.281 ± 0.015 ppm)间无显著差异(P > 0.05),但CO2排放量显著低于河中心沙质沉积物CO2 (500.0 ± 2.3 ppm) (P < 0.01)。16S rRNA基因焦磷酸测序分析发现,河中心沙质沉积物相对丰度>1%的优势反硝化菌只有Flavobacterium (1.65%),而Hydrogenophaga相对丰度仅为0.20%。Pearson相关性分析结果表明,Flavobacterium与pH、沙粒含量和CH4排放量呈极显著正相关关系,与水分及养分含量呈显著负相关关系;Hydrogenophaga与Flavobacterium的情况恰恰相反,与水分及养分含量、N2O和CO2排放量呈现显著正相关关系,与pH、沙粒含量和CH4排放量呈极显著负相关关系(表2)。Zhang等[29] 研究发现Flavobacterium和Pseudomonas存在于同一生境时相互竞争,Flavobacterium比Pseudomonas生长快速会优先占据营养丰富区,而假单胞菌属Pseudomonas由于竞争力弱被束缚在营养耗尽区。这和本研究Hydrogenophaga与Flavobacterium在同一生境下相互竞争的结果有一致性。这说明微生物群落构建(community assembly)过程中由于个体间的竞争,表现出某些个体对特定环境因子的嗜好,即生态位的分化[30] -[32] 。
5. 结论
通过细菌16S rRNA基因焦磷酸测序在锡林河底泥中共检测出20个潜在反硝化细菌属。其中,黄杆菌属Flavobacterium和噬氢菌属Hydrogenophaga 2个优势反硝化菌共同占有的比例高达69.03%,接近反硝化菌群总组成的70%。两者存在生态位的分化:Flavobacterium更喜欢粘附在pH相对高的沙粒上,促进CH4的排放并对N2O和CO2排放产生抑制作用,而Hydrogenophaga更喜欢粘附在营养丰富的粉粒或粘粒上,促进N2O和CO2排放并对CH4的产生有抑制作用。
致 谢
感谢国家自然科学基金项目(No. 41361053)、内蒙古自然科学基金项目(No. 2011MS0603)、国家科技部“973计划”前期研究项目(No. 2009CB125909)和内蒙古大学校级大学生创新计划项目(No. 2012234)在资金方面的资助,感谢上海美吉生物医药科技有限公司在测序和生物信息学分析中提供的服务。
参考文献 (References)
- [1] Calderer, M., Martí, V., de Pablo, J., Guivernau, M., Prenafeta-Boldú, F.X. and Viñas, M. (2014) Effects of enhanced denitrification on hydrodynamics and microbial community structure in a soil column system. Chemosphere, 111, 112-119.
- [2] Chen, Y., Zhou, W., Li, Y., Zhang, J., Zeng, G., Huang, A. and Huang, J. (2014) Nitrite reductase genes as functional markers to investigate diversity of denitrifying bacteria during agricultural waste composting. Applied Microbiology and Biotechnology, 98, 4233-4243.
- [3] Jin, Z., Ji, F.Y., Xu, X., Xu X.Y., Chen, Q.K. and Li, Q. (2014) Microbial and metabolic characterization of a denitrifying phosphorus-uptake/side stream phosphorus removal system for treating domestic sewage. Biodegradation, 25, 777-786.
- [4] Wang, Y., Wiatrowski, H.A., John, R., Lin, C.C., Young, L.Y., Kerkhof, L.J., Yee, N. and Barkay, T. (2013) Impact of mercury on denitrification and denitrifying microbial communities in nitrate enrichments of subsurface sediments. Biodegradation, 24, 33-46.
- [5] Peralta, A.L., Matthews, J.W. and Kent, A.D. (2014) Habitat specialization along a wetland moisture gradient differs between ammonia-oxidizing and denitrifying microorganisms. Microbial Ecology, 68, 339-350.
- [6] Angnes, G., Nicoloso, R.S., da Silva, M.L.B., de Oliveira, P.A.V., Higarashi, M.M., Mezzari, M.P. and Miller, P.R.M. (2013) Correlating denitrifying catabolic genes with N2O and N2 emissions from swine slurry composting. Bioresource Technology, 140C, 368-375.
- [7] Shen, Z., Wang, D., Ruan, Y., Xue, C., Xue, C., Zhang, J., Li, R. and Shen, Q. (2014) Deep 16S rRNA pyrosequencing reveals a bacterial community associated with banana Fusarium wilt disease suppression induced by bio-organic fertilizer application. PLoS ONE, 9, e98420.
- [8] Idris, A., Al-Saleh, M., Piatek, M.J., Al-Shahwan, I., Ali, S. and Brown, J.K. (2014) Viral metagenomics: Analysis of Begomoviruses by illumina high-throughput sequencing. Viruses, 6, 1219-1236.
- [9] Gołębiewski, M., Deja-Sikora, E., Cichosz, M., Tretyn, A. and Wróbel, B. (2014) 16S rDNA pyrosequencing analysis of bacterial community in heavy metals polluted soils. Microbial Ecology, 67, 635-647.
- [10] Zhu, D.C., Tanabe, S.H., Yang, C., Zhang, W.M. and Sun, J.Z. (2013) Bacterial community composition of South China Sea sediments through pyrosequencing-based analysis of 16S rRNA genes. PLoS ONE, 8, e78501.
- [11] 郭丽芸, 时飞, 杨柳燕 (2011) 反硝化菌功能基因及其分子生态学研究进展. 微生物学通报, 38, 583-590.
- [12] Chen, H., Mothapo, N.V. and Shi, W. (2014) The significant contribution of fungi to soil N< sub> 2 O production across diverse ecosystems. Applied Soil Ecology, 73, 70-77.
- [13] 方晶晶, 马传明, 刘存富 (2010) 反硝化细菌研究进展. 环境科学与技术, 33, 206-210, 264.
- [14] Abed, R.M., Lam, P., de Beer, D. and Stief, P. (2013) High rates of denitrification and nitrous oxide emission in arid biological soil crusts from the Sultanate of Oman. International Society for Microbial Ecology, 7, 1862-1875.
- [15] Zhong, L., Du, R., Ding, K., Kang, X., Li, F.Y., Bowatte, S., Hoogendoorn, C.J., Wang, Y., Rui, Y., Jiang, L. and Wang, S. (2014) Effects of grazing on N2O production potential and abundance of nitrifying and denitrifying microbial communities in meadow-steppe grassland in northern China. Soil Biology and Biochemistry, 69, 1-10.
- [16] Guo, G.X., Deng, H., Qiao, M., Yao, H.Y. and Zhu, Y.G. (2013) Effect of long-term wastewater irrigation on potential denitrification and denitrifying communities in soils at the watershed scale. Environmental Science and Technology, 47, 3105-3113.
- [17] Qiu, X.F., Wang, T.W., Zhong, X.M., Du, G.C. and Chen, J. (2012) Screening and characterization of an aerobic nitrifying-denitrifying bacterium from activated sludge. Biotechnology and Bioprocess Engineering, 17, 353-360.
- [18] Qu, J.H., Li, H.F., Yang, J.S. and Yuan, H.L. (2008) Flavobacterium cheniae sp. nov., isolated from sediment of a eutrophic reservoir. International Journal of Systematic and Evolutionary Microbiology, 58, 2186-2190.
- [19] Zhou, M.H., Ye, H.R. and Zhao, X.W. (2014) Isolation and characterization of a novel heterotrophic nitrifying and aerobic denitrifying bacterium Pseudomonas stutzeri KTB for bioremediation of wastewater. Biotechnology and Bioprocess Engineering, 19, 231-238.
- [20] Ishii, S., Ashida, N., Otsuka, S. and Senoo, K. (2010) Isolation of oligotrophic denitrifiers carrying previously uncharacterized functional gene sequences. Applied and Environmental Microbiology, 77, 338-342.
- [21] Li, J.Y., Li, B., Zhou, Y., Xu, J. and Zhao, J. (2011) A rapid DNA extraction method for PCR amplification from wetland soils. Letters in Applied Microbiology, 52, 626-633.
- [22] Schloss, P.D. and Handelsman, J. (2005) Introducing DOTUR, a computer program for defining operational taxonomic units and estimating species richness. Applied and Environmental Microbiology, 71, 1501-1506.
- [23] Park, H.I., Choi, Y.J. and Pak, D. (2005) Autohydrogenotrophic denitrifying microbial community in a glass beads biofilm reactor. Biotechnology Letters, 27, 949-953.
- [24] Michaud, L., Caruso, C., Mangano, S., Interdonato, F., Bruni, V. and Giudice, A.L. (2012) Predominance of Flavobacterium, Pseudomonas, and Polaromonas within the prokaryotic community of freshwater shallow lakes in the northern Victoria Land, East Antarctica. FEMS Microbiology Ecology, 82, 391-404.
- [25] Ye, Z., Wang, F., Bi, H., Wang, Z. and Liu, G.H. (2012) Denitrification of nitrate-contaminated groundwater using a simple immobilized activated sludge bioreactor. Water Science and Technology, 66, 517-524.
- [26] Green, S.J., Prakash, O., Jasrotia, P., Overholt, W.A., Cardenas, E., Hubbard, D., Tiedje, J.M., Watson, D.B., Schadt, C.W., Brooks, S.C., Kostka, J.E., Green, S.J., Prakash, O. and Jasrotia, P. (2012) Denitrifying bacteria from the genus Rhodanobacter dominate bacterial communities in the highly contaminated subsurface of a nuclear legacy waste site. Applied and Environment Microbiology, 78, 1039-1047.
- [27] Green, S.J., Prakash, O., Gihring, T.M., Akob, D.M., Jasrotia, P., Jardine, P.M., Watson, D.B., Brown, S.D., Palumbo, A.V. and Kostka, J.E. (2010) Denitrifying bacteria isolated from terrestrial subsurface sediments exposed to mixedwaste contamination. Applied and Environment Microbiology, 76, 3244-3254.
- [28] Prakash, O., Green, S.J., Jasrotia, P., Overholt, W., Canion, A., Watson, D.B., Brooks, S.C. and Kowalsky, M. (2012) Rhodanobacter denitrificans sp. nov., isolated from nitrate-rich zones of a contaminated aquifer. International Journal of Systematic and Evolutionary Microbiology, 62, 2457-2462.
- [29] Zhang, W., Sileika, T. and Packman, A.I. (2013) Effects of fluid flow conditions on interactions between species in biofilms. FEMS Microbiology Ecology, 84, 344-354.
- [30] 牛克昌, 刘怿宁, 沈泽昊, 何芳良, 方精云 (2009) 群落构建的中性理论和生态位理论. 生物多样性, 17, 579-593.
- [31] Philippot, L., Bru, D., Saby, N.P.A., Čuhel, J., Arrouays, D., Šimek, M. and Hallin, S. (2009) Spatial patterns of bacterial taxa in nature reflect ecological traits of deep branches of the 16S rRNA bacterial tree. Environmental Microbiology, 11, 3096-3104.

NOTES
*通讯作者。
