Bioprocess
Vol.
12
No.
02
(
2022
), Article ID:
52281
,
8
pages
10.12677/BP.2022.122009
基于提高蛋白在内质网中折叠效率的策略促进外源蛋白在毕赤酵母中表达水平的研究进展
韩铭海*,王未鲜,朱国飞,马小彦
贵州理工学院,食品药品制造工程学院,贵州 贵阳
收稿日期:2022年4月29日;录用日期:2022年6月2日;发布日期:2022年6月10日

摘要
新生肽在内质网中的折叠效率对于其在毕赤酵母中的高效表达是至关重要的。在内质网质量控制机制作用下,只有正确折叠的蛋白才能通过内质网通道进入随后的分泌途径,而折叠错误和折叠缓慢的多肽则被剔除出内质网,通过ERAD途径被降解。过表达外源蛋白可能造成内质网内聚集大量的未折叠蛋白,超越内质网的加工能力,从而限制其高水平表达。本文综述了通过共表达分子伴侣或UPR转录因子Hac1p促进外源蛋白表达的研究进展,以期为毕赤酵母蛋白高效表达技术的发展提供借鉴。
关键词
毕赤酵母,内质网胁迫,分子伴侣,非折叠蛋白响应

Research Progress on the Strategy of Improving Protein Folding Efficiency in Endoplasmic Reticulum for Enhancing Heterologous Protein Expression Levels in Pichia pastoris
Minghai Han*, Weixian Wang, Guofei Zhu, Xiaoyan Ma
College of Food and Pharmaceutical Engineering, Guizhou Institute of Technology, Guiyang Guizhou
Received: Apr. 29th, 2022; accepted: Jun. 2nd, 2022; published: Jun. 10th, 2022

ABSTRACT
The efficiency of ER in folding nascent peptides is very important to increased expression of secretory proteins in Pichia pastoris. Under the machinery of quality control, only correctly folded proteins can enter the subsequent secretion pathway through the ER channel, while wrongly or slowly folded peptides will be removed from ER and degraded through ERAD pathway. Over expression of heterologous proteins may lead to the accumulation of large quantities of unfolded peptides, thus exceeding the processing capacity of ER and inhibiting high-level expression of target proteins. This paper reviews the research progress of co-expression of molecular chaperones or UPR transcription factor Hac1p to promote the secretion of heterologous proteins, providing references for the development of techniques for efficient protein expression in Pichia pastoris.
Keywords:Pichia pastoris, ER Stress, Chaperones, The Unfolded Protein Response

Copyright © 2022 by author(s) and Hans Publishers Inc.
This work is licensed under the Creative Commons Attribution International License (CC BY 4.0).
http://creativecommons.org/licenses/by/4.0/


1. 引言
微生物生产的异源蛋白广泛地应用于医药、工业生产和科研领域 [1]。虽然细菌是高效表达异源蛋白的宿主,但是缺乏真核细胞具有的蛋白翻译后修饰(蛋白折叠、糖基化、磷酸化和对新生肽的剪切加工等),这也限制了其应用价值 [1] [2]。而作为真核生物的毕赤酵母(Pichia pastoris,最新分类名为Komagataella pastoris或Komagataella phaffii)具有培养成本低、操作简单、易于诱导和高效的分泌表达蛋白能力等优点 [1],而且,该表达系统具有和高级动物细胞类似的蛋白加工机制和翻译后修饰,使通过其生产的异源蛋白更适用于医药和医学诊断领域 [1] [2],并且已成功地表达了重组疫苗,干扰素,白细胞介素、胰岛素等激素、胰高血糖素、人类生长激素和促红细胞生成素和抗体,显示了其优良的应用前景。毕赤酵母全基因组已经完成测序并公布 [3],这无疑将促进基于基因工程改造进一步优化其作为蛋白表达宿主的性能。
蛋白在成熟和分泌到胞外之前,在分泌途径(The secretory pathway)会经历一系列的翻译后修饰加工过程,例如前导肽的切除、新生肽折叠和糖基化等。该途径较为复杂,涉及数量和种类众多的功能蛋白 [4]。新生肽在内质网的折叠加工过程可能是一个主要的限制其高效表达的瓶颈,并且提高多肽的折叠加工效率已经成为一种促进外源蛋白表达的有效策略 [4] [5]。
2. 蛋白在内质网中的折叠和质量控制机制
内质网(Endoplasmic reticulum, ER)是真核细胞中膜含量最多的细胞器,是蛋白分泌途径的必经通道 [6]。与多肽的合成相比,肽链在ER中的折叠和随后的加工过程是缓慢的。ER是蛋白合成、折叠和翻译后修饰的加工场所,具有种类繁多且浓度较高的分子伴侣、折叠酶、蛋白修饰酶和其他功能蛋白,保障了其对新生肽加工的高效性 [6]。根据作用机制的不同,ER的分子伴侣系统可以分为两类 [6]:1) 经典的分子伴侣(Classical chaperones),主要是Hsp40和Hsp70家族蛋白(酵母细胞中没有Hsp90家族蛋白);2) 凝集素类分子伴侣,主要是钙联蛋白(Calnexin)和钙网蛋白(Calreticulin,酵母没有钙网蛋白),其通过召集其他功能蛋白例如UGGT、ERp57和CypB等特异性地作用于N–糖蛋白的折叠加工过程,同时也鉴别折叠效率低下或是折叠错误的多肽,并剔除出ER。
细胞的ER是个拥挤的多肽加工车间,大量折叠效率低下和未正确折叠多肽的存在严重影响了ER行使正常功能,这些多肽会被引导离开ER并被细胞降解,只有正确折叠的蛋白质才能经过ER通道并进入随后的分泌途径,这就是ER的质量控制机制(Quality control) [6]。被ER剔除的多肽通过两种途径被细胞降解 [7]:1) ERAD途径,非正确折叠多肽引导进入细胞质中,被泛素缀合酶修饰,最终被蛋白酶体彻底消化;2) 引导部分内质网借助溶酶体和液泡发生细胞自噬。
3. 内质网胁迫和非折叠蛋白反应
如果内质网集聚了大量未折叠或错误折叠的多肽,超出了内质网的加工能力,导致内质网胁迫(ER stress),将诱发细胞上调非折叠蛋白反应(The unfolded protein response,简称UPR)通路 [7]。
根据目前的理论模型,酵母细胞的UPR是一种线性信号通路(图1) [7] [8]。调节UPR的最主要功能蛋白是激酶/RNase、Ire1p和转录因子Hac1p。在非应激条件下,Ire1p以单体形式存在,ER伴侣Kar2p与之结合。在内质网胁迫条件下,大量聚集的未折叠蛋白质将招募大量的Kar2p与之结合,使得Ire1p脱离Kar2p。这导致了原本以单体存在形式存在的Ire1p相互聚集,并通过磷酸化反应激活其RNase功能。活化的Ire1p催化了HAC1 mRNA的剪接反应,去除了其内含子。细胞中HAC1 mRNA是组成性表达的,但由于其内含子的存在,阻止了细胞合成相应的Hac1p蛋白产物。而去除内含子的HAC1 mRNA则引导细胞合成Hac1p。最后,Hac1p被转移到细胞核,作为转录因子与UPR反应元件(UPRE)结合,从而作用于UPR下游靶基因 [7] [8]。UPR通路下游靶基因功能涉及内质网对新生肽的折叠加工机制、翻译后修饰、蛋白转运、ERAD途径、细胞能量代谢、细胞器自噬和菌体生长等,对特定蛋白表达的作用十分复杂,表现出了较强的蛋白特异性。激发UPR定向促进蛋白表达技术的关键性问题有待解决,但可以明确的是,在UPR激发下,内质网的分子伴侣、辅助分子伴侣(Co-chaperones)、辅助因子、氧化还原酶、ATP、参加蛋白糖基化修饰的酶类和其他功能蛋白都会显著上调,强化了内质网对新生肽的折叠和修饰加工能力(图1) [7] [8]。
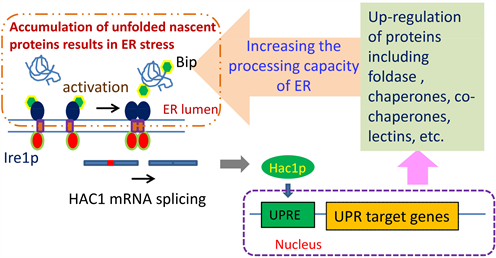
Figure 1. Improving the processing ability of ER for new peptides via activation of UPR [7] [8]
图1. 激发UPR通路提高内质网对新生肽的加工能力 [7] [8]
4. 提高蛋白折叠效率促进毕赤酵母表达外源蛋白
通过提高蛋白在ER中的折叠效率已成为一种提高毕赤酵母外源蛋白表达水平的常用手段,这种策略主要是通过共表达ER分子伴侣和UPR转录因子Hac1p蛋白来实现。
4.1. 共表达分子伴侣PDI
虽然少部分文献报道了共表达二硫键异构酶(PDI)对外源蛋白表达没有显著影响,甚至表现出抑制作用 [9] [10] [11],大部分研究结果还是肯定了共表达PDI促进外源蛋白表达的作用。比如,通过这种策略促进了白介素1受体拮抗剂(融合了人血清白蛋白HSA或人生长激素) [12]、脂肪酶r27RCL (来源于Rhizopuschinensis CCTCC M201021) [13]、碱性嗜冷磷脂酶C [14] 和人类溶菌酶(hLYZ) [15] 的表达。
这种策略对外源蛋白表达的促进作用可能与蛋白本身的二硫键有关。Li等人通过共表达PDI提高了重组家蚕乙酰胆碱酯酶2 (rBmAChE2)的表达水平的5倍。在rBmAChE2蛋白中存在4个二硫键,推断共表达PDI有助于rBmAChE2折叠过程中二硫键的形成,这是促进其表达的重要原因 [16];另外,为了提高AppA植酸酶的热稳定性,Navone等人通过引入二硫键成功地获得了热稳定性更好的突变ApV1,而共表达PDI使ApV1蛋白表达水平提高了12倍 [17]。
Yang等人通过共表达PDI促进了哺乳动物肽聚糖识别蛋白(PGLYRP-1)的表达,而且还发现PDI的基因剂量与PGLYRP-1表达水平的提高密切关联;另外,研究还发现,过表达PDI和Kar2p却抑制了PGLYRP-1的转录水平,他们推断过表达UPR通路的靶基因(PDI和Kar2p)下调了UPR水平,从而抑制了目标蛋白的转录水平 [18]。
另外,目的基因的拷贝数也会影响这种策略的效果。Huang等人研究发现,毕赤酵母表达4拷贝脂肪酶(RML;来源于Rhizomucormiehei)基因时出现了内质网胁迫,而表达2拷贝RML基因则没有这个现象;共表达PDI提高4拷贝RML菌株的表达水平,而对2拷贝RML的菌株却没有作用 [19]。
4.2. 共表达分子伴侣Kar2p
共表达Kar2p对毕赤酵母表达外源蛋白的作用具有较强的蛋白特异性。
共表达Kar2p促进了毕赤酵母表达一些外源蛋白。例如采用这种策略提高了A33轻链抗体片段(A33scFv)表达水平的3倍 [9];提高了人血清白蛋白融合蛋白IL2-HSA表达水平的1.9倍 [20];提高了脂肪酶MAS1表达水平的1.7倍 [21]。同样,共表达Kar2p对表达头孢菌素C酰化酶(SECA) [22] 和疏水蛋白HFBI [23] 也都有促进作用。
然而,这种策略对于某些蛋白的表达却没有效果,甚至表现出了抑制作用。例如,共表达Kar2p对磷脂酶C [10] 和К-卡拉胶酶 [11] 的表达没有影响,反而抑制了南极假丝酵母脂肪酶B [24] 和哺乳动物肽聚糖识别蛋白(PGLYRP-1) [18] 的表达。
根据现有的UPR理论模型,内质网内Kar2p数量与UPR通路水平密切关联 [7] [8]。共表达Kar2p导致内质网内存在高浓度的Kar2p,有足够数量的Kar2p会结合到Ire1p上,阻碍Ire1p相互聚集,使其无法催化HAC1mRNA,这将抑制细胞的UPR水平 [18] [24]。这可以解释共表达Kar2p并没有促进某些外源蛋白的表达,甚至表现了抑制作用。
4.3. 共表达异源的分子伴侣或是折叠酶
有研究表明通过共表达异源的分子伴侣(非来源于毕赤酵母)是一种新的、有效的提高外源蛋白表达水平的策略。例如,Gasser等人通过共表达来源于酿酒酵母的PDI和Kar2p基因分别提高了单克隆抗体片段(2F5mAb)表达水平的1.7和1.5倍 [25]。Summpunn等人在来源于大肠杆菌的分子伴侣GroES-GroEL中融合了毕赤酵母内质网驻留信号序列(HDEL),通过这种技术引导重组GroES-GroEL在毕赤酵母内质网中发挥分子伴侣的作用,研究结果表明这种策略提高了植酸酶表达水平的1.5~2.3倍,但是对于胞内表达的D-苯甘氨酸转氨酶却没有作用 [26]。Huo等人通过共表达人类二硫键异构酶(hPDI)提高了牛卵泡刺激素(bFSH)在毕赤酵母中表达水平的6倍 [27]。而Zhang等人研究发现,共表达同源的折叠酶是成功表达来源于Pseudomonas alcaligenes的脂肪酶必要条件 [28]。
4.4. 激发UPR通路促进外源蛋白表达
4.4.1. 毕赤酵母的HAC1基因
Guerfal等人首次克隆并鉴定了毕赤酵母的HAC1基因,并鉴定了内含子切除位点;研究发现,用UPR诱发剂DTT (二硫苏糖醇)处理细胞,提高了HAC1mRNA丰度;即使在非内质网胁迫条件下,HAC1mRNA都是以去除内含子的活性状态HAC1mRNA(S)存在,并没有发现含有内含子的HAC1mRNA(U) [29]。毕赤酵母具有一定本底UPR水平 [29] [30],这也许可以解释为什么毕赤酵母是良好的蛋白表达宿主。而Whyteside等人却有不同研究报道 [31]:他们检测到了胞内存在HAC1mRNA(U);而用DTT处理细胞后,出现了HAC1mRNA(S),而对照细胞则只有HAC1mRNA(U)。在关于共表达HAC1基因影响外源蛋白表达的文献中,有些并没有明确共表达的是HAC1mRNA(U)还是HAC1mRNA(S)对应的DNA序列;笔者通过共表达HAC1mRNA(S)对应的DNA序列,发现这可以有效地激发毕赤酵母的UPR通路 [32] [33]。
4.4.2. 共表达HAC1基因激发UPR促进蛋白表达
通过共表达UPR下游靶基因的转录因子HAC1基因,可显著地提高细胞UPR水平,这也是目前激发UPR最常用的方法 [5]。虽然,激发UPR对蛋白表达的作用表现出一定的蛋白特异性,但通过这种策略成功提高蛋白表达水平的报道越来越多 [29] [32] - [37]。
共表达HAC1基因的作用与外源蛋白基因拷贝数有关。Lin等人研究发现,共表达HAC1基因对毕赤酵母表达表达单拷贝木聚糖酶A (来源于Bacillus halodurans C-125)没有影响,但能显著促进4拷贝木聚糖酶A的表达 [38];Yang等人也报道了这种策略的效果与哺乳动物肽聚糖识别蛋白(PGLYRP-1)的基因拷贝数量有关联 [18]。
另外,HAC1基因的拷贝数也影响外源蛋白的表达作用,例如Huang等人的研究表明,共表达多拷贝HAC1基因显著促进了α-淀粉酶在毕赤酵母中的表达 [39]。
引导HAC1基因表达的启动子也会影响对外源蛋白的促进作用。例如,Elena等人利用不同转录活性的突变PAOX1启动子引导HAC1基因共表达,结果导致了磷脂酶C表达水平的差异,其中采用30%转录活性(与野生启动子相比较)的突变PAOX1启动子表达HAC1基因时,最利于目的蛋白的表达 [10]。另外,采用启动子PGAP或PAOX1共表达HAC1基因对外源蛋白的表达的效果也不同 [32] [39]。
Bankefa等人 [40] 还研究了共表达不同来源的HAC1基因对表达β-半乳糖苷酶、β-甘露聚糖酶和葡萄糖氧化酶的影响;研究结果表明,PpHac1p (来源于毕赤酵母)、ScHac1p (来源于酿酒酵母)、TrHac1p (来源于里氏木霉)和HsXbp1 (来源于人类)的共表达都有促进外源蛋白表达的可能;不同的外源蛋白最适的Hac1p是不同的,例如,HsXbp1对β-甘露聚糖酶的表达有促进作用,而PpHac1p对此蛋白的表达却没有影响。
4.4.3. 共表达分子伴侣或HAC1基因对细胞生长的影响
值得关注的是,共表达分子伴侣可能影响毕赤酵母的生理特性,比如细胞的生长。比如,共表达HAC1基因会诱发细胞处于高水平UPR状态,导致上调ERAD途径,甚至发生ER自噬现象(细胞剔除部分内质网以缓解内质网胁迫) [41],而且持续的激发UPR状态也会影响细胞的正常生长 [29] [38] [42]。有研究表明共表达PDI会减缓毕赤酵母的生长 [43];Duan等人也报道了共表达Kar2p降低了这种宿主的最高比生长速率 [22]。因此,在考察共表达分子伴侣或HAC1基因对外源蛋白表达的作用时,也必需要考虑宿主细胞生物量的差异对外源蛋白表达的影响。
5. 研究展望
表达分子伴侣或HAC1基因可以缓解由于大量非折叠蛋白聚集造成内质网胁迫,也是提高蛋白在ER中折叠效率的有效手段,但是其生物效应也十分复杂,这种策略对蛋白表达的作用也有一定的蛋白特异性。本文虽然列举了共表达分子伴侣或HAC1基因成功促进蛋白表达的一些案例,但对于特定蛋白,可能需要若干次不同策略的尝试后才能确定较好的技术方案。笔者综合文献并结合个人的研究经验,提出了以下几点建议:1) 某些情况下,只有当毕赤酵母表达高拷贝外源蛋白基因时,共表达分子伴侣或是HAC1基因才有效。表达低拷贝外源蛋白基因时,内质网折叠加工新生肽的效率可能不会成为其高水平表达的瓶颈。2) 尝试共表达不同来源(非毕赤酵母)的分子伴侣或是HAC1基因可能会获得更好的效果。3) 尝试采用强弱不同的启动子来表达分子伴侣或是HAC1基因也是有意义的。高水平而持续地表达分子伴侣或HAC1基因可能会打破内质网工作机制和分泌途径的平衡,有时不一定有利于蛋白的表达。4) 要充分考虑外源蛋白本身的背景。比如外源蛋白含有较多的二硫键,那么共表达PDI可能会有较好的效果;一般来说,蛋白本身的热稳定性与其折叠的难易程度有关 [44],如果外源蛋白具有较强的热稳定性,那么这种蛋白在内质网中的折叠效率可能就比较高,为了提高其表达水平,首先要尝试增加目的蛋白基因拷贝数,然后再考虑共表达分子伴侣或HAC1基因的策略。
基金项目
本文受到国家自然科学基金项目(No. 31760022)、贵州省科技计划项目(黔科合基础[2020]1Y148号)和贵州理工学院高层次人才科研启动项目(XJGC20190626)的资助。
文章引用
韩铭海,王未鲜,朱国飞,马小彦. 基于提高蛋白在内质网中折叠效率的策略促进外源蛋白在毕赤酵母中表达水平的研究进展
Research Progress on the Strategy of Improving Protein Folding Efficiency in Endoplasmic Reticulum for Enhancing Heterologous Protein Expression Levels in Pichia pastoris[J]. 生物过程, 2022, 12(02): 81-88. https://doi.org/10.12677/BP.2022.122009
参考文献
- 1. Karbalaei, M., Rezaee, S.A. and Farsiani, H. (2020) Pichia pastoris: A Highly Successful Expression System for Opti-mal Synthesis of Heterologous Proteins. Journal of Cellular Physiology, 235, 5867-5881. https://doi.org/10.1002/jcp.29583
- 2. Baghban, R., Farajnia, S., Rajabibazl, M., et al. (2019) Yeast Expression Systems: Overview and Recent Advances. Molecular Biotechnology, 61, 365-384. https://doi.org/10.1007/s12033-019-00164-8
- 3. De Schutter, K., Lin, Y.C., Tiels, P., et al. (2009) Genome Se-quence of the Recombinant Protein Production Host Pichia pastoris. Nature Biotechnology, 27, 561-566. https://doi.org/10.1038/nbt.1544
- 4. Puxbaum, V., Mattanovich, D. and Gasser, B. (2015) Quo Vadis? The Chal-lenges of Recombinant Protein Folding and Secretion in Pichia pastoris. Applied Microbiology and Biotechnology, 99, 2925-2938. https://doi.org/10.1007/s00253-015-6470-z
- 5. Yu, P., Zhu, Q., Chen, K., et al. (2015) Improving the Secretory Production of the Heterologous Protein in Pichia pastoris by Focusing on Protein Folding. Applied Biochemistry and Bi-otechnology, 175, 535-548. https://doi.org/10.1007/s12010-014-1292-5
- 6. Araki, K. and Nagata, K. (2012) Protein Folding and Quality Control in the ER. Cold Spring Harbor Perspectives in Biology, 4, a015438. https://doi.org/10.1101/cshperspect.a015438
- 7. Read, A. and Schröder, M. (2021) The Unfolded Protein Re-sponse: An Overview. Biology, 10, Article No. 384. https://doi.org/10.3390/biology10050384
- 8. Snapp, E.L. (2012) Unfolded Protein Responses with or without Unfolded Proteins? Cells, 1, 926-950. https://doi.org/10.3390/cells1040926
- 9. Damasceno, L.M., Pla, I., Chang, H.J., et al. (2004) An Optimized Fer-mentation Process for High-Level Production of a Single-Chain Fv Antibody Fragment in Pichia pastoris. Protein Ex-pression and Purification, 37, 18-26. https://doi.org/10.1016/j.pep.2004.03.019
- 10. Elena, C., Ravasi, P., Cerminati, S., et al. (2016) Pichia pastoris Engineering for the Production of a Modified Phospholipase C. Process Biochemistry, 51, 1935-1944. https://doi.org/10.1016/j.procbio.2016.08.022
- 11. Yu, Y., Liu, Z., Chen, M., et al. (2020) Enhancing the Expres-sion of Recombinant κ-Carrageenase in Pichia pastoris Using Dual Promoters, Co-Expressing Chaperones and Tran-scription Factors. Biocatalysis and Biotransformation, 38, 104-113. https://doi.org/10.1080/10242422.2019.1655001
- 12. Shen, Q., Wu, M., Wang, H.B., et al. (2012) The Effect of Gene Copy Number and Co-Expression of Chaperone on Production of Albumin Fusion Proteins in Pichia pastoris. Ap-plied Microbiology and Biotechnology, 96, 763-772. https://doi.org/10.1007/s00253-012-4337-0
- 13. Sha, C., Yu, X.W., Lin, N.X., et al. (2013) Enhancement of Li-pase r27RCL Production in Pichia pastoris by Regulating Gene Dosage and Co-Expression with Chaperone Protein Di-sulfide Isomerase. Enzyme and Microbial Technology, 53, 438-443. https://doi.org/10.1016/j.enzmictec.2013.09.009
- 14. Wang, L., Hu, T., Jiang, Z., et al. (2021) Efficient Production of a Novel Alkaline Cold-Active Phospholipase C from Aspergillus Oryzaeby Molecular Chaperon Co-Expression for Crude Oil Degumming. Food Chemistry, 350, Article ID: 129212. https://doi.org/10.1016/j.foodchem.2021.129212
- 15. He, H., Wu, S., Mei, M., et al. (2020) A Combinational Strategy for Effective Heterologous Production of Functional Human Lysozyme in Pichia Pastoris. Frontiers in Bioen-gineering and Biotechnology, 8, Article No. 118. https://doi.org/10.3389/fbioe.2020.00118
- 16. Li, J., Cai, J., Ma, M., et al. (2021) Preparation of a Bombyx mori Acetylcholinesterase Enzyme Reagent through Chaperone Protein Disulfide Isomerase Co-Expression Strategy in Pichia pastoris for Detection of Pesticides. Enzyme and Microbial Technology, 144, Article ID: 109741. https://doi.org/10.1016/j.enzmictec.2020.109741
- 17. Navone, L., Vogl, T., Luangthongkam, P., et al. (2021) Di-sulfide Bond Engineering of AppA Phytase for Increased Thermostability Requires Co-Expression of Protein Disulfide Isomerase in Pichia pastoris. Biotechnology for Biofuels and Bioproducts, 14, 80. https://doi.org/10.1186/s13068-021-01936-8
- 18. Yang, J., Lu, Z., Chen, J., et al. (2016) Effect of Cooperation of Chaperones and Gene Dosage on the Expression of Porcine PGLYRP-1 in Pichia pastoris. Applied Microbiology and Biotechnology, 100, 5453-5465. https://doi.org/10.1007/s00253-016-7372-4
- 19. Huang, J., Zhao, Q., Chen, L., et al. (2020) Improved Production of Recombinant Rhizomucor miehei Lipase by Coexpressing Protein Folding Chaperones in Pichia pastoris, Which Triggered ER Stress. Bioengineered, 11, 375-385. https://doi.org/10.1080/21655979.2020.1738127
- 20. Guan, B., Chen, F., Su, S., et al. (2016) Effects of Co-Overexpression of Secretion Helper Factors on the Secretion of a HSA Fusion Protein (IL2-HSA) in Pichia pastoris. Yeast, 33, 587-600. https://doi.org/10.1002/yea.3183
- 21. Lan, D., Qu, M., Yang, B., et al. (2016) Enhancing Pro-duction of Lipase MAS1 from Marine Streptomyces sp. Strain in Pichia pastoris by Chaperones Co-Expression. Elec-tronic Journal of Biotechnology, 22, 62-67. https://doi.org/10.1016/j.ejbt.2016.06.003
- 22. Duan, G., Ding, L., Wei, D., et al. (2019) Screening Endogenous Signal Peptides and Protein Folding Factors to Promote the Secretory Expression of Heterologous Proteins in Pichia pastoris. Journal of Biotechnology, 306, 193-202. https://doi.org/10.1016/j.jbiotec.2019.06.297
- 23. Sallada, N.D., Harkins, L.E. and Berger, B.W. (2019) Effect of Gene Copy Number and Chaperone Coexpression on Recombinant Hydrophobin HFBI Biosurfactant Production in Pichia pastoris. Biotechnology and Bioengineering, 116, 2029-2040. https://doi.org/10.1002/bit.26982
- 24. Samuel, P., Prasanna Vadhana, A.K., Kamatchi, R., et al. (2013) Effect of Molecular Chaperones on the Expression of Candida antarctica Lipase B in Pichia pastoris. Microbiological Research, 168, 615-620. https://doi.org/10.1016/j.micres.2013.06.007
- 25. Gasser, B., Sauer, M., Maurer, M., et al. (2007) Transcriptom-ics-Based Identification of Novel Factors Enhancing Heterologous Protein Secretion in Yeasts. Applied and Environ-mental Microbiology, 73, 6499-6507. https://doi.org/10.1128/AEM.01196-07
- 26. Summpunn, P., Jomrit, J. and Panbangred, W. (2018) Improvement of Extracellular Bacterial Protein Production in Pichia pastoris by Co-Expression of Endoplasmic Reticulum Residing GroEL-GroES. Journal of Bioscience and Bioengineering, 125, 268-274. https://doi.org/10.1016/j.jbiosc.2017.09.007
- 27. Huo, X., Liu, Y., Wang, X., et al. (2007) Co-Expression of Hu-man Protein Disulfide Isomerase (hPDI) Enhances Secretion of Bovine Follicle-stimulating Hormone (bFSH) in Pichia pastoris. Protein Expression and Purification, 54, 234-239. https://doi.org/10.1016/j.pep.2007.03.016
- 28. Zhang, Z., Zhang, X., Hao, H., et al. (2020) Co-Expression of Pseudomonas alcaligenes Lipase and Its Specific Foldase in Pichia pastoris by a Dual Expression Cassette Strategy. Protein Expression and Purification, 175, Article ID: 105721. https://doi.org/10.1016/j.pep.2020.105721
- 29. Guerfal, M., Ryckaert, S., Jacobs, P.P., et al. (2010) The HAC1 Gene from Pichia pastoris: Characterization and Effect of Its Overexpression on the Production of Secreted, Surface Displayed and Membrane Proteins. Microbial Cell Factories, 9, Article No. 49. https://doi.org/10.1186/1475-2859-9-49
- 30. Fauzee, Y., Taniguchi, N., Ishiwata-Kimata, Y., et al. (2020) The Unfolded Protein Response in Pichia pastoris without External Stressing Stimuli. FEMS Yeast Research, 20, foaa053. https://doi.org/10.1093/femsyr/foaa053
- 31. Whyteside, G., Nor, R.M., Alcocer, M.J., et al. (2011) Activation of the Unfolded Protein Response in Pichia pastoris Requires Splicing of a HAC1 mRNA Intron and Retention of the C-Terminal Tail of Hac1p. FEBS Letters, 585, 1037-1041. https://doi.org/10.1016/j.febslet.2011.02.036
- 32. Han, M., Wang, W., Gong, X., et al. (2021) Increased Expression of Recombinant Chitosanase by Co-Expression of Hac1p in the Yeast Pichia pastoris. Protein and Peptide Letters, 28, 1434-1441. https://doi.org/10.2174/0929866528666211105111155
- 33. Han, M., Wang, W., Zhou, J., et al. (2020) Activation of the Unfolded Protein Response via Co-Expression of the HAC1i Gene Enhances Expression of Recombinant Elastase in Pichia pastoris. Biotechnology and Bioprocess Engineering, 25, 302-307. https://doi.org/10.1007/s12257-019-0381-2
- 34. Wang, Y., Luo, X., Zhao, Y., et al. (2021) Integrated Strategies for Enhancing the Expression of the AqCoA Chitosanase in Pichia pastoris by Combined Optimization of Molecular Chaperones Combinations and Copy Numbers via a Novel Plasmid pMC-GAP. Applied Biochemistry and Biotechnology, 193, 4035-4051. https://doi.org/10.1007/s12010-021-03668-9
- 35. De Waele, S., Vandenberghe, I., Laukens, B., et al. (2018) Op-timized Expression of the Starmerella bombicola Lactone Esterase in Pichia pastoris through Temperature Adaptation, Codon-Optimization and Co-Expression with HAC1. Protein Expression and Purification, 143, 62-70. https://doi.org/10.1016/j.pep.2017.10.016
- 36. Gasser, B., Maurer, M., Gach, J., et al. (2006) Engineering of Pichia pastoris for Improved Production of Antibody Fragments. Biotechnology and Bioengineering, 94, 353-361. https://doi.org/10.1002/bit.20851
- 37. Krainer, F.W., Gerstmann, M.A., Darnhofer, B., et al. (2016) Biotechnolog-ical Advances towards an Enhanced Peroxidase Production in Pichia pastoris. Journal of Biotechnology, 233, 181-189. https://doi.org/10.1016/j.jbiotec.2016.07.012
- 38. Lin, X.Q., Liang, S.L., Han, S.Y., et al. (2013) Quantitative iTRAQ LC-MS/MS Proteomics Reveals the Cellular Response to Heterologous Protein Overexpression and the Regula-tion of HAC1 in Pichia pastoris. Journal of Proteomics, 2013, 91, 58-72. https://doi.org/10.1016/j.jprot.2013.06.031
- 39. Huang, M., Gao, Y., Zhou, X., et al.(2017) Regulating Unfolded Protein Response Activator HAC1p for Production of Thermostable Raw-Starch Hydrolyzing α-Amylase in Pichia pas-toris. Bioprocess and Biosystems Engineering, 40, 341-350. https://doi.org/10.1007/s00449-016-1701-y
- 40. Bankefa, O.E., Wang, M., Zhu, T., et al. (2018) Hac1p Homo-logues from Higher Eukaryotes Can Improve the Secretion of Heterologous Proteins in the Yeast Pichia pastoris. Bio-technology Letters, 40, 1149-1156. https://doi.org/10.1007/s10529-018-2571-y
- 41. Kruse, K.B., Brodsky, J.L. and McCracken, A.A. (2006) Au-tophagy: An ER Protein Quality Control Process. Autophagy, 2, 135-137. https://doi.org/10.4161/auto.2.2.2388
- 42. Liu, J., Han, Q., Cheng, Q., et al. (2020) Efficient Expression of Human Lysozyme through the Increased Gene Dosage and Co-Expression of Transcription Factor Hac1p in Pichia pastoris. Current Microbiology, 77, 846-854. https://doi.org/10.1007/s00284-019-01872-9
- 43. Liu, X., Wu, D., Wu, J., et al. (2013) Optimization of the Pro-duction of Aspergillus niger α-Glucosidase Expressed in Pichia pastoris. World Journal of Microbiology & Biotechnol-ogy, 29, 533-540. https://doi.org/10.1007/s11274-012-1207-y
- 44. Whyteside, G., Alcocer, M.J., Kumita, J.R., et al. (2011) Na-tive-State Stability Determines the Extent of Degradation Relative to Secretion of Protein Variants from Pichia pastoris. PLOS ONE, 6, e22692. https://doi.org/10.1371/journal.pone.0022692
NOTES
*通讯作者。