Advances in Clinical Medicine
Vol.
13
No.
04
(
2023
), Article ID:
63903
,
12
pages
10.12677/ACM.2023.134771
基于GEO数据库构建神经母细胞瘤缺氧相关的预后模型
李苏曼,鹿洪亭*
青岛大学附属青岛市妇女儿童医院小儿外科,山东 青岛
收稿日期:2023年3月9日;录用日期:2023年4月5日;发布日期:2023年4月14日

摘要
目的:基于基因表达综合(Gene Expression Omnibus, GEO)数据库分析缺氧相关基因与神经母细胞瘤(NB)临床特征及免疫环境的关系,构建缺氧相关预后模型。方法:从GEO数据库下载数据集GSE62564,从MSigDB数据库下载缺氧相关基因集。通过共识聚类算法将NB患者分为高低缺氧组,通过Kaplan-Meier曲线、卡方检验、“ESTIMATE”算法等比较两组间临床、免疫环境差别。通过LASSO回归建立缺氧相关预后模型,使用受试者工作特征(ROC)曲线下面积(AUC)评价模型的预测性能,并对模型进行独立预后分析。结果:根据缺氧相关基因将NB样本分为两组,两组患者的生存、年龄、肿瘤分期及MYCN状态均存在显著差异。两组样本的免疫浸润情况存在显著差异,B细胞、树突状细胞、NK细胞及T细胞等多种免疫细胞含量在两组间存在差异。通过LASSO回归得到了6个缺氧相关基因(CSRP2, DTNA, SAP30, NCAN, WSB1, NAGK)构成的预后模型。通过模型的风险评分中位数将患者分为高、低风险组,Kaplan-Meier曲线显示两组间预后存在显著差异。模型在1,3,5年时的AUC值分别为0.882,0.916及0.914,说明其具有良好的预测价值。多因素COX回归分析表明风险评分可作为NB的独立预后因素。结论:缺氧影响NB的免疫环境及患者预后,由缺氧相关基因构成的预后模型能较好评估NB患者预后,并为寻找新的治疗靶点提供帮助。
关键词
神经母细胞瘤,缺氧,预后模型,GEO数据库

Construction of Hypoxia-Related Prognostic Signature in Neuroblastoma Based on GEO Database
Suman Li, Hongting Lu*
Department of Pediatric Surgery, Qingdao Women and Children’s Hospital Affiliated to Qingdao University, Qingdao Shandong
Received: Mar. 9th, 2023; accepted: Apr. 5th, 2023; published: Apr. 14th, 2023

ABSTRACT
Aims: Analyzing relationship between hypoxia-related genes and clinical features and immune environment of neuroblastoma (NB) based on GEO (Gene Expression Omnibus) database to construct hypoxia-related prognostic signature. Methods: GSE62564 was downloaded from GEO database and hypoxia-related gene set was obtained from MSigDB database. Patients in GSE62564 were divided into high/low hypoxia subgroups via Consensus Clustering analysis. Differences in immune and clinical features between two groups were identified by Kaplan-Meier curve, Chi-square test and ESTIMATE algorithm. Hypoxia-related prognostic signature was constructed by Lasso-Cox regression. AUC value of the signature was calculated for the evaluation of the prognostic model. Results: Clinical features including prognosis, age, tumor stage and MYCN status were significantly different in two subgroups. Immune environment and distribution of immune cell including B cells, T cells, NK cells and dendritic cells were also different between groups. A six-gene hypoxia-related prognostic signature (CSRP2, DTNA, SAP30, NCAN, WSB1, NAGK) was constructed using Lasso-Cox regression, and patients were divided into high-/low-risk group by median of risk score. Kaplan-Meier curve revealed the significant difference between prognosis of the two groups. The AUCs for the 1-, 3-, 5-year OS predictions for the signature were 0.882, 0.916 and 0.914 which revealed the prognostic value of the signature. Risk score was identified as an independent prognostic factor by the multivariate Cox regression analysis. Conclusion: Hypoxia has an effect on immune environment and prognosis of NB patients. A hypoxia-related prognostic signature was constructed, which will contribute to predicting prognosis of NB and finding new therapy target.
Keywords:Neuroblastoma, Hypoxia, Prognostic Signature, Gene Expression Omnibus Database

Copyright © 2023 by author(s) and Hans Publishers Inc.
This work is licensed under the Creative Commons Attribution International License (CC BY 4.0).
http://creativecommons.org/licenses/by/4.0/


1. 引言
神经母细胞瘤(neuroblastoma, NB)是一种交感神经系统颅外实体瘤,源自神经嵴细胞。作为最常见的儿童实体肿瘤,90%的NB发生于5岁之前 [1] 。因NB死亡的人数约占儿童癌症相关死亡人数的10% [2] 。NB的临床表现具有很强的异质性。部分NB患者可自发缓解,而另一部分患者即使经过手术治疗,肿瘤仍会发生进展 [3] 。根据MYCN状态、年龄、分期及分级,可将NB患者分为高风险及低风险级别,例如Susan等在2009年的研究中描述的国际神经母细胞瘤风险组(INRG) [4] 。尽管手术、放化疗和免疫治疗等治疗手段在不断进步,高风险患者的五年生存率仍低于60% [5] [6] 。因此,寻找新的生物标志物和治疗靶点有重要的临床意义。缺氧是实体肿瘤的重要特征。肿瘤细胞快速增殖及肿瘤血管结构、功能异常都会导致肿瘤组织区域缺氧的发生 [7] 。在包括NB在内的多种肿瘤中,缺氧状态都被证实可以影响肿瘤的进展、侵袭和转移 [8] [9] [10] 。此外,肿瘤的缺氧状态与肿瘤免疫微环境也可发生相互作用 [11] 。本研究通过生物信息学分析缺氧相关基因与NB临床特征及免疫环境的关系,并构建缺氧相关预后模型,为NB的早期诊断与治疗提供新思路。
2. 资料与方法
2.1. 数据收集与处理
从基因表达综合(Gene Expression Omnibus, GEO)数据库下载数据集GSE62564。从分子特征数据库(Molecular Signatures Database, MSigDB)下载缺氧相关数据集,数据集内包含200个缺氧相关基因。
2.2. 缺氧相关基因的共识聚类分析
在R软件版本(4.3.0)中使用“ConsensuClusterPlus”包对GSE62564中的样本进行聚类分析。利用缺氧相关基因的表达量将样本分为两组。研究中使用了1000次迭代的共识聚类算法,在每次迭代中采样80%的数据。使用“survival”包对两组患者的预后情况进行分析并绘制Kaplan-Meier曲线。
2.3. 缺氧状态与肿瘤免疫环境的相关性分析
使用R软件中的“CIBERSORT”通过反卷积算法,估算肿瘤样本中22种免疫细胞的组成比例,并比较不同分组间免疫细胞比例差异。通过“Estimate”算法计算每个样本的免疫评分及间质评分,比较高、低缺氧分组间分数的差异。
2.4. 缺氧相关基因预后风险模型的构建与评价
下载GSE62564数据集中患者的生存信息,使用R中的“survival”包探究缺氧相关基因对NB患者预后的影响。P小于0.05的基因作为生存相关基因。使用“glmnet”包,基于LASSO回归算法建立缺氧相关预后模型,并通过预后模型为每个患者计算风险评分。接着,患者根据风险评分的中位数被分为高风险和低风险组。通过Kaplan-Meier曲线比较两组间预后情况。通过R中的“ROCR”包构建模型的受试者工作(receiver operating characteristic, ROC)曲线并计算曲线下面积(area under curve, AUC)值。最后通过单因素、多因素分析验证风险评分是否为独立预后因素。
2.5. 列线图模型的构建
使用R包“rms”,综合风险评分及年龄、临床分期、MYCN状态及肿瘤进展情况,构建列线图预后模型。使用“ROCR”包绘制该模型用于预测NB患者1,3,5年生存情况的ROC曲线。使用“rmda”包绘制临床决策曲线(DCA)。
3. 结果
3.1. 通过缺氧相关基因对NB进行分类
下载GSE62564数据集中498位患者的mRNA表达矩阵及临床信息。将从MSigDB数据库中下载的200个缺氧相关基因与GSE62564中的基因名重叠,共有184个缺氧相关基因。通过共识聚类算法,我们将498例NB样本根据缺氧相关基因表达量进行分组。当聚类系数k取2至6时,k = 2时取得较好结果,即组内相关性高,组间相关性低(图1(a))。据此,我们将498例样本分为聚类1 (cluster1)和聚类2 (cluster2),分别包含342例和156例样本。缺氧相关基因在两组间的表达情况如图1(b)所示。Kaplan-Meier生存曲线显示cluster1中患者的预后显著好于cluster2 (P < 0.001),表明缺氧相关基因与患者预后显著相关(图1(c))。
3.2. 不同缺氧状态下的免疫环境探究
肿瘤缺氧状态与肿瘤免疫微环境关系密切。为探究不同缺氧状态下肿瘤微环境的改变,我们使用R中的“ESTIMATE”算法评估不同样本免疫浸润的状况。如图2(a)所示,cluster1中样本的基质评分及免疫评分均高于cluster2,表明cluster1中样本具有更多的免疫细胞及基质细胞浸润。为进一步探究各样本中免疫细胞浸润的具体情况,我们使用“CIBERSORT”算法评估22种免疫细胞在样本中的含量。如图2(b)所示,B细胞、树突状细胞、NK细胞及T细胞等多种免疫细胞含量在两组间存在差异。
3.3. 探究缺氧状态与临床特征的关系
Cluster1及cluster2中患者的临床信息如表1所示,包括年龄、性别、临床分期及MYCN状态。使用卡方检验比较两组患者的临床特征。结果显示,两组患者的年龄、临床分期、MYCN状态存在显著差异(P < 0.05)。两组患者的性别无显著差异。

Figure 1. Dividing NB patients into high/low hypoxia groups by hypoxia-related genes
图1. 通过缺氧相关基因将NB患者分为高/低缺氧组
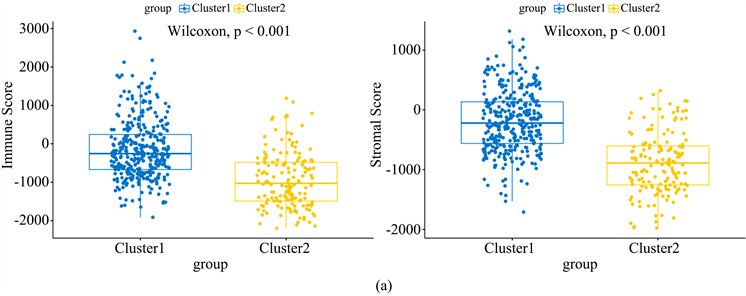

Figure 2. Differences in immune environment between cluster 1 and cluster 2
图2. 组1和组2间免疫环境的差别
Table 1. Relationship between hypoxia and clinical features of NB patients
表1. 缺氧与NB患者临床特征的相关性
3.4. 构建缺氧相关预后模型
为进一步完善对NB患者预后的评估,并筛选可能的治疗靶点,我们构建了缺氧相关预后模型。首先,通过单因素COX分析方法,结合498位患者的预后信息,筛选对NB患者生存具有显著影响的缺氧相关基因,共106个(P < 0.05)。将这106个生存相关基因纳入分析,通过LASSO回归算法构建预后模型(图3(a))。构建的预后模型由六个缺氧相关基因(CSRP2, DTNA, SAP30, NCAN, WSB1, NAGK)及相应系数组成。通过预后模型为每个患者计算风险评分,计算公式为:风险评分 = (CSRP2表达量 × 0.08370066) + (SAP30表达量 × 0.28005528) + (NCAN表达量 × 0.01927312) − (WSB1表达量 × 0.42357840) − (NAGK表达量 × 0.10539461) − (DTNA表达量 × 0.06373091)。
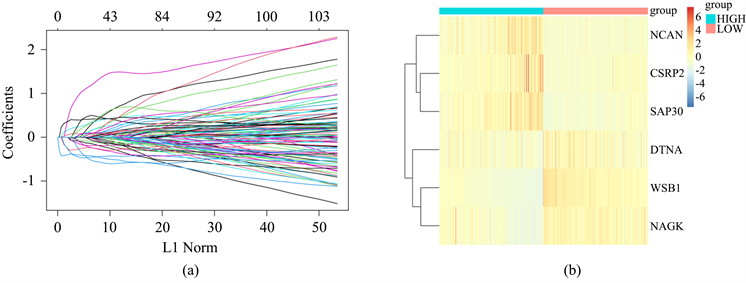
Figure 3. Construction of hypoxia-related prognostic signature by Lasso-Cox regression
图3. 通过LASSO回归构建缺氧相关预后模型
3.5. 对预测模型预测效果的评估
根据风险评分的中位数,将498位患者分为高风险组及低风险组。使用R中的“pheatmap”包绘制6个基因在各样本中的表达情况(图3(b))。通过“survival”包绘制Kaplan-Meier曲线(图4(a)),发现低风险组患者预后显著好于高风险组(P < 0.001)。箱线图(图4(b))说明生存患者的风险评分显著高于死亡患者(P < 0.001)。通过ROC曲线计算预测模型在预测患者1、3、5年时生存情况的AUC值,分别为0.882,0.916及0.914,说明该模型对患者预后具有较好的预测效果(图4(c))。
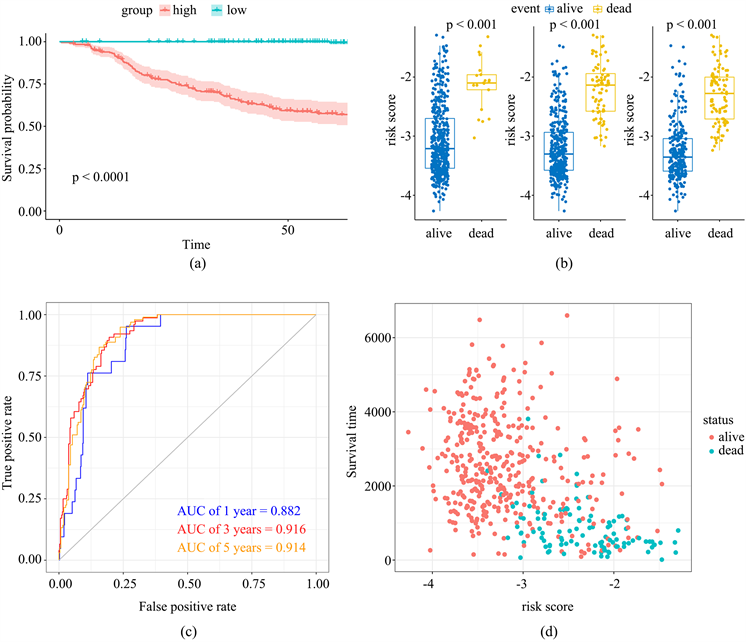
Figure 4. Prediction value of hypoxia-related prognostic signature
图4. 缺氧相关预后模型的预测价值
3.6. 临床指标及风险评分与预后的关系
收集NB患者的临床信息,包括性别、年龄、肿瘤分期、MYCN状态。使用多因素COX回归分析各临床指标及风险评分与预后的关系。如表2所示,年龄、分期、MYCN状态及风险评分与预后显著相关。多因素COX分析表明年龄、分期及风险评分可作为患者预后的独立预测指标。
3.7. 构建列线图模型
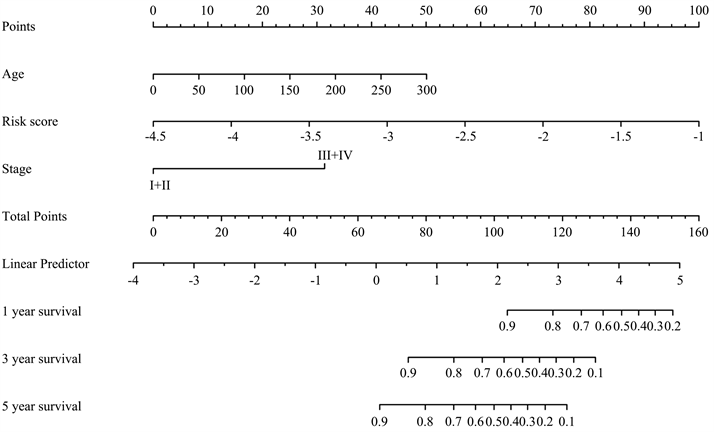
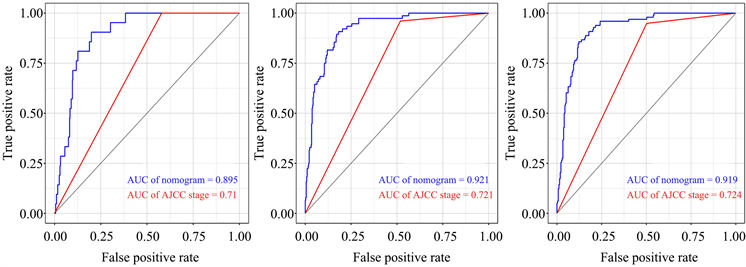
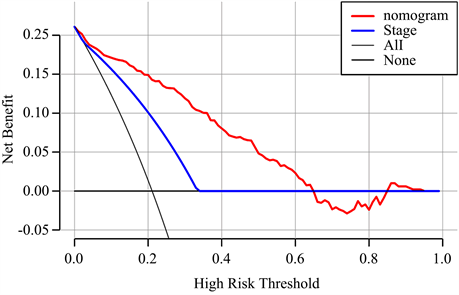
结合风险评分及与预后相关的年龄、肿瘤分期,构建用于预测NB患者预后的列线图模型(图5(a))。根据模型计算每位患者的评分,并通过ROC曲线计算模型在1、3、5年的AUC值,分别为0.895,0.921及0.919。比较列线图与肿瘤分期的AUC值,可见列线图的预测效率高于肿瘤分期(图5(b))。通过临床决策曲线比较列线图模型与肿瘤分期诊断的准确性(图5(c))。
Table 2. The multivariate Cox regression analysis of prognostic factors in NB patients
表2. 影响NB患者预后的多因素分析
 (a)
(a)
 (b)
(b)
 (c)
(c)
Figure 5. Construction of prognostic nomogram
图5. 构建列线图模型
4. 讨论
神经母细胞瘤(NB)是一种在交感神经系统发育过程中,起源于神经嵴的实体肿瘤 [12] 。尽管NB的发病率并不高,但其仍是儿童最常见的颅外恶性实体肿瘤,占据儿童癌症死亡人数的10% [13] 。NB具有很强的异质性,其中高风险亚型对传统治疗反应差,患者预后较差 [14] 。肿瘤细胞增殖具有快速且不受控制的特点,限制了肿瘤区域氧气的使用。因此,血液供应不足及缺氧是几乎所有实体肿瘤的微环境特征 [15] 。虽然在肿瘤中有新生血管形成,但持续缺氧会引起肿瘤血管分布不规则,毛细血管之间距离增加,超过氧气的扩散能力 [16] [17] 。肿瘤细胞在缺氧环境中侵袭性及耐药性可能增强。缺氧诱导基因表达变化,对各种细胞的生理功能有重要影响,并最终影响患者预后 [18] 。例如在肝细胞癌中,缺氧可通过影响Treg细胞及树突状细胞,介导肝细胞癌免疫抑制 [19] 。在乳腺癌中,缺氧相关基因ADAM12的表达变化可介导肿瘤的侵袭与转移 [20] 。
作为明确的肿瘤特征,已有研究关注缺氧及缺氧相关基因在NB发生、发展中的作用。但缺氧状态对于NB免疫环境及患者预后的影响有待进一步探究。本研究通过共识聚类算法,依据MSigDB数据库中的缺氧相关基因集将GSE62564中的498位患者分为高、低缺氧分组,并比较两组间临床特征及免疫环境的区别。本研究中,不同缺氧状态的患者,其免疫微环境有显著差异。接着,使用LASSO回归算法建立了由CSRP2,DTNA,SAP30,NCAN,WSB1,NAGK六个缺氧相关基因组成的预后风险模型。该模型具有良好的预测性能,并可作为NB的独立预后因素。最后,将风险评分与临床信息结合,构建列线图模型,进一步提升预测效率。
本预后模型由6个缺氧相关基因组成。CSRP2属于富含半胱氨酸和甘氨酸蛋白(cysteine and glycine rich protein, CSRP)家族,编码LIM结构域蛋白,参与调节发育和细胞分化过程。CSRP2在多种肿瘤中起调控作用。在乳腺癌中,HIF-1可通过靶向CSRP2,促进乳腺癌细胞的侵袭 [21] 。而在结肠癌中,CSRP2可通过HIPPO信号通路抑制肿瘤细胞增殖,且CSRP2的低表达与结肠癌患者的不良预后相关 [22] 。DNTA主要编码肌营养不良蛋白相关蛋白复合物(dystrophin-associated protein complex, DAPC)中的支架蛋白,主要作用是维持心肌、骨骼肌的结构完整性 [23] 。Hu等 [24] 在研究中发现,DNTA可通过调控STAT3、TGFβ1和P53信号通路促进乙肝病毒相关肝癌的进展。在食管癌中,microRNA-301b-DNTA轴可影响肿瘤细胞的生长、侵袭和迁移 [25] 。SAP30编码的蛋白是组蛋白脱乙酰酶复合物的组成成分,已被报道可通过UHRF1-SAP30-MXD4轴影响急性髓性白血病的发展 [26] 。Tang等 [27] 的研究建立了肝细胞癌缺氧相关预后模型,其中SAP30可作为肝细胞癌不良预后的标志物。这与本研究中SAP30的趋势相同。神经蛋白聚糖(neurocan, NCAN)主要参与调节细胞的粘附和迁移。在NB中,NCAN可促进肿瘤细胞的增殖与迁移,并与NB患者的不良预后显著相关 [28] 。此外,NCAN也可在肝等非神经组织中表达,并可能是酒精性肝病引起肝癌的风险因素 [29] 。此前多项关于NB预后模型的研究也将NCAN纳入其中,说明其对NB预后的影响有较大的意义,值得进一步研究 [30] [31] 。NAGK (N-acetylglucosamine kinase)属于乙酰己糖胺激酶家族。在NB患者中,血浆MYCN与NAGK的比值可用于检测MYCN的扩增情况,并可用于判断肿瘤负荷及预后情况 [32] [33] 。此外,NAGK可通过干预细胞内糖代谢影响胰腺癌肿瘤生长及患者预后 [34] [35] 。WSB1属于E3泛素连接酶,参与了多种细胞过程,如甲状腺激素调节、免疫调节和调节细胞对缺氧的反应等 [36] 。据研究,WSB1可通过调控ATM激酶降解来促进肿瘤进展 [37] 。WSB1可通过介导pVHL降解促进肿瘤转移。这在Poujade等关于乳腺癌的研究中得到证实 [38] [39] 。据Keren Shichrur等 [40] 的研究,沉默WSB1可抑制NB细胞的生长。但Chen等 [41] 的研究中指出,WSB1的高表达与较好的临床结局相关,这与本研究相符。因此,WSB1在NB中的作用有待进一步研究。
综上所述,本研究基于GEO数据库,分析了缺氧状态与NB免疫环境及临床特征的关系,并构建了由6个缺氧相关基因组成的预后风险模型,以期更好地判断NB患者的预后,并为NB的治疗提供新的思路。
基金项目
山东省自然科学基金资助项目(No. ZR2020MH213)。
文章引用
李苏曼,鹿洪亭. 基于GEO数据库构建神经母细胞瘤缺氧相关的预后模型
Construction of Hypoxia-Related Prognostic Signature in Neuroblastoma Based on GEO Database[J]. 临床医学进展, 2023, 13(04): 5444-5455. https://doi.org/10.12677/ACM.2023.134771
参考文献
- 1. Anderson, J., Majzner, R.G. and Sondel, P.M. (2022) Immunotherapy of Neuroblastoma: Facts and Hopes. Clinical Cancer Research, 28, 3196-3206. https://doi.org/10.1158/1078-0432.CCR-21-1356
- 2. Smith, M.A., Altekruse. S.F., Adamson. P.C., Reaman, G.H. and Seibel, N.L. (2014) Declining Childhood and Adolescent Cancer Mortality. Cancer, 120, 2497-2506. https://doi.org/10.1002/cncr.28748
- 3. Hains, A.E., Uppal, S., Cao, J.Z., Salwen, H.R., Applebaum, M.A., Cohn, S.L., et al. (2022) MYCN and HIF-1 Directly Regulate TET1 Expression to Control 5-hmC Gains and Enhance Neuroblastoma Cell Migration in Hypoxia. Epigenetics, 17, 2056-2074. https://doi.org/10.1080/15592294.2022.2106078
- 4. Cohn, S.L., Pearson, A.D., London, W.B., Monclair, T., Ambros, P.F., Brodeur, G.M., et al. (2009) The International Neuroblastoma Risk Group (INRG) Classification System: An INRG Task Force Report. Journal of Clinical Oncology, 27, 289-297. https://doi.org/10.1200/JCO.2008.16.6785
- 5. Moreno, L., Guo, D., Irwin, M.S., Berthold, F., Hogarty, M., Ka-mijo, T., et al. (2021) A Nomogram of Clinical and Biologic Factors to Predict Survival in Children Newly Diagnosed with High-Risk Neuroblastoma: An International Neuroblastoma Risk Group Project. Pediatric Blood & Cancer, 68, e28794. https://doi.org/10.1002/pbc.28794
- 6. Bosse, K.R. and Maris, J.M. (2016) Advances in the Translational Genomics of Neuroblastoma: From Improving Risk Stratification and Revealing Novel Biology to Identifying Actionable Genomic Alterations. Cancer, 122, 20-33. https://doi.org/10.1002/cncr.29706
- 7. Vaupel, P., Kelleher, D.K. and Thews, O. (1998) Modulation of Tumor Oxygenation. International Journal of Radiation Oncology, Biology, Physics, 42, 843-838. https://doi.org/10.1016/S0360-3016(98)00324-1
- 8. Curro, M., Ferlazzo, N., Giunta, M.L., Montalto, A.S., Rus-so, T., Arena, S., et al. (2020) Hypoxia-Dependent Expression of TG2 Isoforms in Neuroblastoma Cells as Consequence of Different MYCN Amplification Status. International Journal of Molecular Sciences, 21, Article 1364. https://doi.org/10.3390/ijms21041364
- 9. Bao, M.H. and Wong, C.C. (2021) Hypoxia, Metabolic Reprogram-ming, and Drug Resistance in Liver Cancer. Cells, 10, Article 1715. https://doi.org/10.3390/cells10071715
- 10. Hao, S., Zhu, X., Liu, Z., Wu, X., Li, S., Jiang, P., et al. (2021) Chron-ic Intermittent Hypoxia Promoted Lung Cancer Stem Cell-Like Properties via Enhancing Bach1 Expression. Respiratory Research, 22, Article No. 58. https://doi.org/10.1186/s12931-021-01655-6
- 11. Jing, X., Yang, F., Shao, C., Wei, K., Xie, M., Shen, H., et al. (2019) Role of Hypoxia in Cancer Therapy by Regulating the Tumor Microenvironment. Molecular Cancer, 18, Article No.157. https://doi.org/10.1186/s12943-019-1089-9
- 12. Marshall, G.M., Carter, D.R., Cheung, B.B., Liu, T., Mateos, M.K., Meyerowitz, J.G., et al. (2014) The Prenatal Origins of Cancer. Nature Reviews Cancer, 14, 277-289. https://doi.org/10.1038/nrc3679
- 13. Smith, M.A., Seibel, N.L., Altekruse, S.F., Ries, L.A., Melbert, D.L., O’Leary, M., et al. (2010) Outcomes for Children and Adolescents with Cancer: Challenges for the Twenty-First Century. Journal of Clinical Oncology, 28, 2625-2634. https://doi.org/10.1200/JCO.2009.27.0421
- 14. Matthay, K.K., Maris, J.M., Schleiermacher, G., Nakagawara, A., Mackall, C.L., Diller, L., et al. (2016) Neuroblastoma. Nature Reviews Disease Primers, 2, Article No. 16078. https://doi.org/10.1038/nrdp.2016.78
- 15. Shao, C., Yang, F., Miao, S., Liu, W., Wang, C., Shu, Y., et al. (2018) Role of Hypoxia-Induced Exosomes in Tumor Biology. Molecular Cancer, 17, Article No. 120. https://doi.org/10.1186/s12943-018-0869-y
- 16. Wigerup, C., Påhlman, S. and Bexell, D. (2016) Therapeutic Tar-geting of Hypoxia and Hypoxia-Inducible Factors in Cancer. Pharmacology & Therapeutics, 164, 152-169. https://doi.org/10.1016/j.pharmthera.2016.04.009
- 17. Wilson, W.R. and Hay, M.P. (2011) Targeting Hypoxia in Cancer Therapy. Nature Reviews Cancer, 11, 393-410. https://doi.org/10.1038/nrc3064
- 18. Roma-Rodrigues, C., Mendes, R., Baptista, P.V. and Fernandes, A.R. (2019) Targeting Tumor Microenvironment for Cancer Therapy. International Journal of Molecular Sciences, 20, Article 840. https://doi.org/10.3390/ijms20040840
- 19. Suthen, S., Lim, C.J., Nguyen, P.H.D., Dutertre, C.A., Lai, H.L.H., Wasser, M., et al. (2022) Hypoxia-Driven Immunosuppression by Treg and Type-2 Conventional Dendritic Cells in HCC. Hepatology, 76, 1329-1344. https://doi.org/10.1002/hep.32419
- 20. Wang, R., Godet, I., Yang, Y., Salman, S., Lu, H., Lyu, Y., et al. (2021) Hypoxia-Inducible Factor-Dependent ADAM12 Expression Mediates Breast Cancer Invasion and Metastasis. Proceed-ings of the National Academy of Sciences, 118, e2020490118.https://doi.org/10.1073/pnas.2020490118
- 21. Hoffmann, C., Mao, X., Brown-Clay, J., Moreau, F., Al Absi, A., Wurzer, H., et al. (2018) Hypoxia Promotes Breast Cancer Cell Invasion through HIF-1α-Mediated Up-Regulation of the Invadopodial Actin Bundling Protein CSRP2. Scientific Reports, 8, Article No. 10191. https://doi.org/10.1038/s41598-018-28637-x
- 22. Chen, L., Long, X., Duan, S., Liu, X., Chen, J., Lan, J., et al. (2020) CSRP2 Suppresses Colorectal Cancer Progression via p130Cas/Rac1 Axis-Meditated ERK, PAK, and HIPPO Signaling Pathways. Theranostics, 10, 11063-11079. https://doi.org/10.7150/thno.45674
- 23. Cao, Q., Shen, Y., Liu, X., Yu, X., Yuan, P., Wan, R., et al. (2017) Phe-notype and Functional Analyses in a Transgenic Mouse Model of Left Ventricular Noncompaction Caused by a DTNA Mutation. International Heart Journal, 58, 939-947. https://doi.org/10.1536/ihj.16-019
- 24. Hu, Z.G., Zhang, S., Chen, Y.B., Cao, W., Zhou, Z.Y., Zhang, J.N., et al. (2020) DTNA Promotes HBV-Induced Hepatocellular Carcinoma Progression by Activating STAT3 and Regulating TGFbeta1 and P53 Signaling. Life Sciences, 258, Article 118029. https://doi.org/10.1016/j.lfs.2020.118029
- 25. Fu, G., Pei, Z. and Song, N. (2021) Oncogenic microRNA-301b Regulates Tumor Repressor Dystrobrevin Alpha to Facilitate Cell Growth, Invasion and Migration in Esophageal Cancer. Esophagus, 18, 315-325. https://doi.org/10.1007/s10388-020-00764-3
- 26. Hu, C.L., Chen, B.Y., Li, Z., Yang, T., Xu, C.H., Yang, R., et al. (2022) Targeting UHRF1-SAP30-MXD4 Axis for Leukemia Initiating Cell Eradication in Myeloid Leukemia. Cell Re-search, 32, 1105-1123. https://doi.org/10.1038/s41422-022-00735-6
- 27. Tang, Y., Zhang, H., Chen, L., Zhang, T., Xu, N. and Huang, Z. (2022) Identification of Hypoxia-Related Prognostic Signature and Competing Endogenous RNA Regulatory Axes in Hepatocellular Carcinoma. International Journal of Molecular Sciences, 23, Article 13590. https://doi.org/10.3390/ijms232113590
- 28. Su, Z., Kishida, S., Tsubota, S., Sakamoto, K., Cao, D., Kiyonari, S., et al. (2017) Neurocan, an Extracellular Chondroitin Sulfate Proteoglycan, Stimulates Neuroblastoma Cells to Promote Malignant Phenotypes. Oncotarget, 8, 106296- 106310. https://doi.org/10.18632/oncotarget.22435
- 29. Nischalke, H.D., Lutz, P., Krämer, B., Söhne, J., Müller, T., Rosendahl, J., et al. (2014) A Common Polymorphism in the NCAN Gene Is Associated with Hepatocellular Carcinoma in Alcoholic Liver Disease. Journal of Hepatology, 61, 1073-1079. https://doi.org/10.1016/j.jhep.2014.06.006
- 30. Zhong, X., Liu, Y., Liu, H., Zhang, Y., Wang, L. and Zhang, H. (2018) Identification of Potential Prognostic Genes for Neuroblastoma. Frontiers in Genetics, 9, Article 589. https://doi.org/10.3389/fgene.2018.00589
- 31. Tan, K., Wu, W., Zhu, K., Lu, L. and Lv, Z. (2022) Identification and Characterization of a Glucometabolic Prognostic Gene Signature in Neuroblastoma Based on N6-Methyladenosine Eraser ALKBH5. Journal of Cancer, 13, 2105-2125. https://doi.org/10.7150/jca.69408
- 32. Su, Y., Wang, L., Zhao, Q., Yue, Z., Zhao, W., Wang, X., et al. (2020) Im-plementation of the Plasma MYCN/NAGK Ratio to Detect MYCN Amplification in Patients with Neuroblastoma. Molecu-lar Oncology, 14, 2884-2893. https://doi.org/10.1002/1878-0261.12794
- 33. Shirai, R., Osumi, T., Sato-Otsubo, A., Nakabayashi, K., Ishiwata, K., Yamada, Y., et al. (2022) Quantitative Assessment of Copy Number Alterations by Liquid Biopsy for Neuroblastoma. Genes, Chromosomes and Cancer, 61, 662- 669. https://doi.org/10.1002/gcc.23073
- 34. Campbell, S., Mesaros, C., Izzo, L., Affronti, H., Noji, M., Schaffer, B.E., et al. (2021) Glutamine Deprivation Triggers NAGK-Dependent Hexosamine Salvage. Elife, 10, e62644. https://doi.org/10.7554/eLife.62644
- 35. Tang, H., Wei, P., Chang, P., Li, Y., Yan, D., Liu, C., et al. (2017) Genetic Polymorphisms Associated with Pancreatic Cancer Survival: A Genome-Wide Association Study. International Journal of Cancer, 141, 678-686. https://doi.org/10.1002/ijc.30762
- 36. Haque, M., Kendal, J.K., MacIsaac, R.M. and Demetrick, D.J. (2016) WSB1: From Homeostasis to Hypoxia. Journal of Biomedical Science, 23, Article No. 61. https://doi.org/10.1186/s12929-016-0270-3
- 37. Kim, J.J., Lee, S.B., Yi, S.Y., Han, S.A., Kim, S.H., Lee, J.M., et al. (2017) WSB1 Overcomes Oncogene-Induced Senescence by Targeting ATM for Degradation. Cell Research, 27, 274-293. https://doi.org/10.1038/cr.2016.148
- 38. Kim, J.J., Lee, S.B., Jang, J., Yi, S.Y., Kim, S.H., Han, S.A., et al. (2015) WSB1 Promotes Tumor Metastasis by Inducing pVHL Degradation. Genes & Development, 29, 2244-2257. https://doi.org/10.1101/gad.268128.115
- 39. Poujade, F.A., Mannion, A., Brittain, N., Theodosi, A., Beeby, E., Leszczynska, K.B., et al. (2018) WSB-1 Regulates the Metastatic Potential of Hormone Receptor Negative Breast Cancer. British Journal of Cancer, 118, 1229-1237. https://doi.org/10.1038/s41416-018-0056-3
- 40. Shichrur, K., Feinberg-Gorenshtein, G., Luria, D., Ash, S., Yaniv, I. and Avigad, S. (2014) Potential Role of WSB1 Isoforms in Growth and Survival of Neuroblastoma Cells. Pediatric Research, 75, 482-486. https://doi.org/10.1038/pr.2014.2
- 41. Chen, Q.R., Bilke, S., Wei, J.S., Greer, B.T., Steinberg, S.M., Westermann, F., et al. (2006) Increased WSB1 Copy Number Correlates with Its Over-Expression Which Associates with Increased Survival in Neuroblastoma. Genes, Chromosomes and Cancer, 45, 856-862. https://doi.org/10.1002/gcc.20349
NOTES
*通讯作者。
