Hans Journal of Biomedicine
Vol.08 No.02(2018), Article ID:24330,11
pages
10.12677/HJBM.2018.82003
Protein-Protein-Interaction Network and Pathway Analysis Related to Alzheimer’s Disease
Yuchen Xu1, Lin Wei2, Honglei Li3
1Department of Neurology, Xiangya Hospital, Central South University, Changsha Hunan
2Xiangya School of Medicine, Central South University, Changsha Hunan
3Department of Otorhinolaryngology, Third Xiangya Hospital, Central South University, Changsha Hunan

Received: Mar. 21st, 2018; accepted: Apr. 2nd, 2018; published: Apr. 9th, 2018

ABSTRACT
Alzheimer's disease (AD) is one of the most common neurodegenerative disease, and its pathogenesis is not fully understood. This article derives transcriptome data of patients with AD from the Gene Expression database GEO (Gene Expression Omnibus), through which we obtain the AD-related risk gene set. Based on AD risk gene set, we construct a protein-protein-interaction network of AD. At the same time we used functional enrichment analysis and path analysis to explore the biological processes and signal pathway involved in AD. Our work helps to elucidate the pathogenic mechanism of AD and provide guidance to the future prevention and treatment of AD.
Keywords:Alzheimer’s Disease, Protein-Protein-Interaction Network, Functional Enrichment Analysis
阿尔兹海默症发病相关蛋白互作网络构建 与通路分析
徐煜宸1,魏琳2,李泓磊3
1中南大学湘雅医院神经内科,湖南 长沙
2中南大学湘雅医学院,湖南 长沙
3中南大学湘雅三医院耳鼻喉科,湖南 长沙

收稿日期:2018年3月21日;录用日期:2018年4月2日;发布日期:2018年4月9日

摘 要
阿兹海默症(Alzheimer’s diseas, AD)是最常见的神经退行性疾病,其发病机制尚未完全明确。本文拟从基因表达综合数据库GEO (Gene Expression Omnibus)数据库中获取大样本量的AD患者的转录组数据,通过分析得到AD风险基因集,以AD风险基因集为基础,构建AD相关风险基因的蛋白互作网络发掘枢纽基因。同时利用功能富集分析和通路分析探索AD发病过程中涉及的生物学过程和信号通路,从而为阐明阿尔兹海默病的致病机制和探索阿尔兹海默病的防治新思路提供指导。
关键词 :阿尔兹海默症,蛋白互作网络,功能富集分析

Copyright © 2018 by authors and Hans Publishers Inc.
This work is licensed under the Creative Commons Attribution International License (CC BY).
http://creativecommons.org/licenses/by/4.0/


1. 引言
阿尔兹海默病(Alzheimer’s disease, AD,以下简称AD)是一种最常见的痴呆症。在1906年,德国精神病医生和神经病理学家Alois Alzheimer首次报道了这种疾病的症状和特征 [1] 。患者随着病情推进逐渐表现出渐进性记忆障碍、认知功能障碍、人格改变以及语言障碍等神经精神症状 [2] [3] [4] 。随着老年人占社会人口比例的上升,AD患者的总数和每年新患病人数会以较快的速度增长。此疾病目前还不能被治愈,且患者生活质量极为低下,需要他人长期护理,这给患者家庭与整个社会带来了沉重的精神压力和经济负担。AD相关研究因其艰巨性和重要的社会效应而显得愈发重要。目前学界已经普遍承认遗传因素在AD的发病中起到了至关重要的作用 [5] [6] 。家族型AD大多由以下三个基因突变引起:APP、PSNE1和PSNE2 [7] 。Lambert等人利用全基因组关联分析发现19个基因影响AD的发病率 [8] 。但现有的证据尚不足以全面且透彻的阐释AD的发病机理。
随着对人类疾病研究的不断深入,学界开始认可,许多复杂疾病的发生常常不是单一基因的突变而引起的,其发病过程往往有多个基因的参与,基因与基因之间互相作用,相互影响,形成网络 [9] 。从多基因共同作用引起疾病发生的角度来研究疾病发生机制和遗传基础,是更准确的更有效的方法。以复数个基因组成的疾病风险基因集进行复杂疾病的深入研究已经成为研究疾病发病机制的一个重要手段。
本文通过GEO数据库所获取的大样本量AD转录本数据,得到AD相关的风险基因集,构建AD相关风险基因的蛋白互作网络发掘枢纽基因。同时利用功能富集分析和通路分析探索AD发病过程中涉及的生物学过程和信号通路,对AD进行系统生物学层面的生物信息学分析,为AD的预防和治疗探索新思路。
2. 方法
2.1. AD相关风险基因集的获取
以GEO数据库为基础,检索策略为:
(((“alzheimer disease” [MeSH Terms] OR Alzheimer’s disease [All Fields]) AND brain [Sample Source]) NOT ((“micrornas” [MeSH Terms] OR micrornas [All Fields]) OR (“micrornas” [MeSH Terms] OR miRNA [All Fields]) OR (“rna, untranslated” [MeSH Terms] OR non coding RNA [All Fields]) OR (small [All Fields] AND (“rna” [MeSH Terms] OR RNA[All Fields])))) AND “Homo sapiens” [porgn] AND “GSE” [Filter]。限定时间范围为1950-1-1至2018-1-5,最终共获得结果27条。
纳入标准为:
1) 样本为人类大脑;
2) 有至少5例AD患者样本。
3) 存在合理的健康对照样本。
排除标准为:
1) 基因甲基化的相关研究。
2) 不提供原始数据的研究。
3) 不提供样本信息的研究。
4) 研究非编码RNA的文章。
5) 样本量过小、数据残缺不全的研究。
最终符合要求数据源共6个(GSE1297, GSE28146, GSE39420, GSE4757, GSE5281, GSE48350)。
使用R语言进行数据预处理:数据过滤和数据转换、处理缺失数据和批次校正。
通过BH算法,以矫正p值小于0.05为标准,得到各数据源的差异表达基因集。为确保结果的可信度,将六个数据源得到的基因集取交集,最终得到本文使用的AD相关风险基因集。
2.2. 蛋白互相作用网络的构建
蛋白互相作用背景网络来自于人类蛋白质文献数据库(Human Protein Reference Database, HPRD),其所有数据信息均为学者从已发表文献中分析而来,其中包含了蛋白的结构域、翻译后修饰、相互作用等相关信息,具有可靠性较高。本研究通过该数据库网站下载其中蛋白互相作用关系部分的所有信息,用于构建蛋白互相作用预测模型 [10] 。
2.3. AD相关风险基因的功能富集分析
基因富集分析和基因本体论(Gene Ontology, GO)功能注释采用DAVID(The Database for Annotation, Visualization and Integrated Discovery, DAVID, version 6.7)分析。DAVID数据库是一个基于web的工具,可以提供高通量数据集(诸如表达谱和蛋白组平台)的注释和分析的整合解决方案 [11] 。本文以PBH < 0.05为界,限定显著富集的生物学过程。
人类AD风险通路数据的富集分析数据来源于京都基因与基因组百科全书(Kyoto Encyclopedia of Genes and Genomes, KEGG) [12] 。
3. 结果
3.1. AD风险基因集
六个数据源的差异表达基因取交集后最后得到76个基因纳入AD风险基因集(表1)。
3.2. AD风险基因互相作用网络
去除无互相作用关系的孤立节点,得到一个存在49个节点(风险基因),249项互作关系的互相作用网络(图1)。
3.3. 功能富集分析
利用在线工具DAVID,本文从生物学过程(biological process, BP),分子功能(Molecular function, MF)
Table 1. AD-related gene set
表1. AD风险基因集
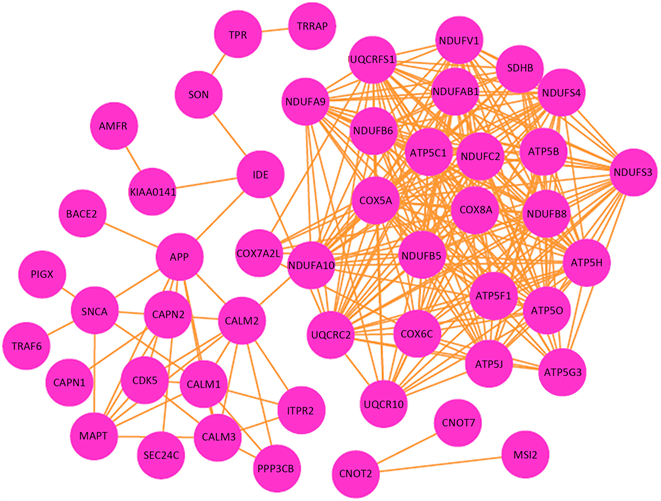
Figure 1. Network of AD related genes
图1. AD风险基因互相作用网络
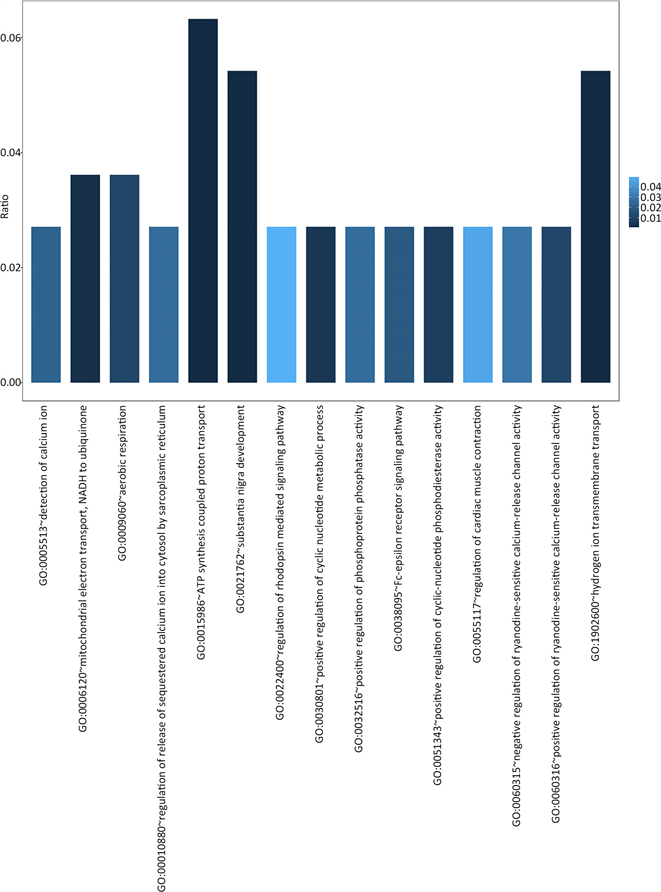
Figure 2. Biological process GO terms significantly enriched in AD related genes
图2. 显著富集于AD风险基因集的BP GO terms
和细胞组分(Cellular component, CC)三个范畴对76个AD风险基因进行了GO富集分析。判定最终结果有意义的标准如下:
1) 多重检验校正后的p值即PBH < 0.05,
2) 每条GO术语(terms)中至少包含3个AD风险基因。
共有38个GO terms满足上述条件,其中BP有15个(图2),MF有9个(图3),CC有14个(图4)。
3.4. 通路分析
识别候选基因集中的富集通路,将为我们理解AD发病机制进一步提供重要信息。
判定最终结果有意义的标准如下:
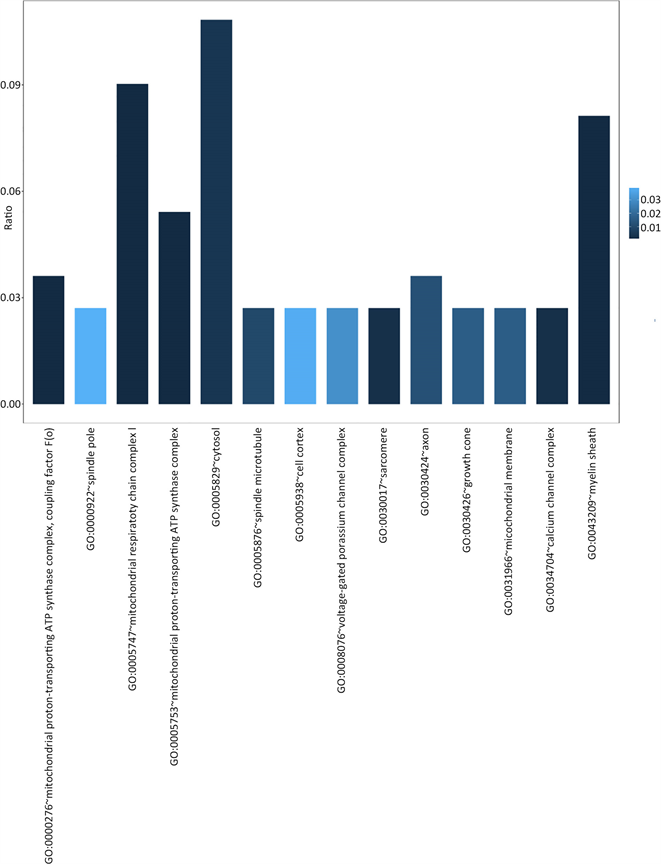
Figure 3. Cellular component GO terms significantly enriched in AD related genes
图3. 显著富集于AD风险基因集的细胞组分GO terms

Figure 4. Molecular function GO terms significantly enriched in AD related genes
图4. 显著富集于AD风险基因集的分子功能GO terms
1) 多重检验校正后的p值即PBH < 0.05。
2) 每条通路中至少包含5个AD风险基因。
共有7条通路符合要求,在表2中展示。
4. 讨论
AD是当前最为普遍的神经退行性疾病之一,在既往的几十年中,其发病机制的研究在基因层面上主
Table 2. Significantly enriched pathway in AD related genes
表2. AD相关基因的显著富集通路
要着眼于明确的家系 [13] [14] ,对于散发AD尚缺乏一个较广维度的探索,在分子层面上则主要集中在淀粉样蛋白(β-amyiod)和Tau蛋白上 [15] [16] [17] [18] ,目前还很少有研究从分子互作网络的角度探索AD的潜在分子机制。本文首次从大样本量的AD转录组数据出发,获取AD发病相关的风险基因,并构建互相作用网络,从系统生物学层面,分析AD在发病过程中可能涉及的生物学过程和信号通路。
通过对6个AD患者脑组织转录组数据的处理分析,并取差异表达基因的交集后,我们得到了一个包含76个基因的AD风险基因集,值得一提的是,经过文献检索,其中35个在既往的研究中,被确切的实验验证与AD的发病存在关系,具体见表3。此结果一方面佐证了我们所获得的76个AD风险基因是确切可信的,另一方面也高度提示另外41个尚未见报道的基因极有可能也在AD的发病中发挥着重要的作用,值得未来实验的探索。
构建蛋白质互相作用网络后我们发现,仅有一部分基因的产物存在互相作用关系,相比那些孤立的基因,与其他基因存在互相作用关系的基因更可能积极地参与到AD的发生发展中。并且其中网络连接度权重(degree)高的基因很可能起着枢纽基因的作用,作为潜在的上游,调控整个网络并最终促使疾病的发生。在本文构建的网络中,共有49个节点,即49个风险基因。其中,权重最高的三个为ATP5C1,NDUFC2,COX8A。这三个基因的产物分别是ATP合酶成分,NADH脱氢酶辅酶成分,细胞色素c氧化酶成分,这说明能量代谢和氧化应激可能是AD发病的枢纽环节。
通过对AD基因进行GO分析结果,我们可以发现,76个AD风险基因主要涉及的生物学过程和细胞组分包括:能量代谢相关(NADH脱氢酶活性,ATP合成酶,线粒体呼吸链组分等),氧化应激相关(细胞色素C氧化酶成分,胞内氧化还原过程等),离子通道和信号传递相关(电压门控钾离子通道复合体,氢离子跨膜转运活动,钙离子通道复合体等)。这可以进一步说明,AD发生发展的过程与能量代谢的异常以及氧化应激导致的损伤密切相关,而对离子通道功能的损害则是导致AD患者认知功能障碍的重要因素。
通过通路富集分析,我们得到7条显著富集的通路,其中包含基因最多的为KEGG定义的阿尔兹海默症基础疾病通路,这再次佐证了我们获取的AD风险基因集的可信度。其次,许多基因富集与氧化磷酸化和代谢相关通路上,再次说明其在AD发病过程中的重要性。同时我们发现十分有趣的一点,同为神经退行性疾病的帕金森病和亨廷顿病的通路也在富集通路之列,这说明这三种疾病之间可能存在某些相同或相似的生物学过程,这非常值得进一步的探索。
总而言之,本文首次通过系统生物学方法,定义了一组有较高可信度的AD风险基因,并建立了其产物的相互作用网络,通过进一步的生物信息学分析,探索了AD发生发展过程中涉及的生物学过程。
Table 3. Genes related to AD validated by experiments
表3. 被实验验证与AD相关的基因 [19] - [26]
本文的结论,对于进一步探索AD的发病机制和预防治疗策略有一定的指导意义。在未来,在本文的研究基础上,还能继续添加诸如microRNA,lncRNA等更多组学数据,进一步完善互相作用网络。同时,对于本文发现的关键基因,关键通路,十分值得进一步的动物实验及细胞学实验验证。
文章引用
徐煜宸,魏 琳,李泓磊. 阿尔兹海默症发病相关蛋白互作网络构建与通路分析
Protein-Protein-Interaction Network and Pathway Analysis Related to Alzheimer’s Disease[J]. 生物医学, 2018, 08(02): 16-26. https://doi.org/10.12677/HJBM.2018.82003
参考文献
- 1. Apostolova, L.G. (2016) Alzheimer Disease. Continuum, 22, 419-434. https://doi.org/10.1212/CON.0000000000000307
- 2. Balthazar, M.L., Yasuda, C.L., Cendes, F. and Damasceno, B.P. (2010) Learning, Retrieval, and Recognition Are Compromised in aMCI and Mild AD: Are Distinct Episodic Memory Processes Mediated by the Same Anatomical Structures? Journal of the International Neuropsychological Society, 16, 205-209. https://doi.org/10.1017/S1355617709990956
- 3. Barnes, J. and Fox, N.C. (2014) The Search for Early Markers of AD: Hippocampal Atrophy and Memory Deficits. International Psychogeriatrics, 26, 1065-1066. https://doi.org/10.1017/S1041610214000623
- 4. Yang, F.M., Grigorenko, A., Tommet, D., Farias, S.T., Mungas, D., Bennett, D.A., Jones, R.N. and Crane, P.K. (2013) AD Pathology and Cerebral Infarctions Are Associated with Memory and Executive Functioning One and Five Years Before Death. Journal of Clinical and Experimental Neuropsychology, 35, 24-34. https://doi.org/10.1080/13803395.2012.740001
- 5. Bergem, A.L., Engedal, K. and Kringlen, E. (1997) The Role of Heredity in Late-Onset Alzheimer Disease and Vascular Dementia. A Twin Study. Archives of General Psychiatry, 54, 264-270. https://doi.org/10.1001/archpsyc.1997.01830150090013
- 6. Pedersen, N.L., Berg, S., Johansson, B., Johansson, K., Viitanen, M., Winblad, B. and Gatz, M. (1998) Genetic Factors Are Often Found Inl Alzheimer Disease. An Extensive Twin Study to Clarify the Heredity-Environment Relationship. Lakartidningen, 95, 2585-2588.
- 7. Lanoiselee, H.M., Nicolas, G., Wallon, D., Rovelet-Lecrux, A., Lacour, M., Rousseau, S., Richard, A.C., Pasquier, F., Rollin-Sillaire, A., Martinaud, O., Quillard-Muraine, M., de la Sayette, V., Boutoleau-Bretonniere, C., Etcharry-Bouyx, F., Chauvire, V., Sarazin, M., le Ber, I., Epelbaum, S., Jonveaux, T., Rouaud, O., Ceccaldi, M., Felician, O., Godefroy, O., Formaglio, M., Croisile, B., Auriacombe, S., Chamard, L., Vincent, J.L., Sauvee, M., Marelli-Tosi, C., Gabelle, A., Ozsancak, C., Pariente, J., Paquet, C., Hannequin, D., Campion, D. and Collaborators of the CNRMAJp (2017) APP, PSEN1, and PSEN2 Mutations in Early-Onset Alzheimer Disease: A Genetic Screening Study of Familial and Sporadic Cases. PLOS Medicine, 14, e1002270. https://doi.org/10.1371/journal.pmed.1002270
- 8. Lambert, J.C., Ibrahim-Verbaas, C.A., Harold, D., Naj, A.C., Sims, R., Bellenguez, C., DeStafano, A.L., Bis, J.C., Beecham, G.W., Grenier-Boley, B., Russo, G., Thorton-Wells, T.A., Jones, N., Smith, A.V., Chouraki, V., Thomas, C., Ikram, M.A., Zelenika, D., Vardarajan, B.N., Kamatani, Y., Lin, C.F., Gerrish, A., Schmidt, H., Kunkle, B., Dunstan, M.L., Ruiz, A., Bihoreau, M.T., Choi, S.H., Reitz, C., Pasquier, F., Cruchaga, C., Craig, D., Amin, N., Berr, C., Lopez, O.L., De Jager, P.L., Deramecourt, V., Johnston, J.A., Evans, D., Lovestone, S., Letenneur, L., Moron, F.J., Rubinsztein, D.C., Eiriksdottir, G., Sleegers, K., Goate, A.M., Fievet, N., Huentelman, M.W., Gill, M., Brown, K., Kamboh, M.I., Keller, L., Barberger-Gateau, P., McGuiness, B., Larson, E.B., Green, R., Myers, A.J., Dufouil, C., Todd, S., Wallon, D., Love, S., Rogaeva, E., Gallacher, J., St George-Hyslop, P., Clarimon, J., Lleo, A., Bayer, A., Tsuang, D.W., Yu, L., Tsolaki, M., Bossu, P., Spalletta, G., Proitsi, P., Collinge, J., Sorbi, S., Sanchez-Garcia, F., Fox, N.C., Hardy, J., Deniz Naranjo, M.C., Bosco, P., Clarke, R., Brayne, C., Galimberti, D., Mancuso, M., Matthews, F., European Alzheimer’s Disease Initiative, Genetic and Environmental Risk in Alzheimer’s Disease, Alzheimer’s Disease Genetic Consortium, Cohorts for Heart and Aging Research in Genomic Epidemiology, Moebus, S., Mecocci, P., Del Zompo, M., Maier, W., Hampel, H., Pilotto, A., Bullido, M., Panza, F., Caffarra, P., Nacmias, B., Gilbert, J.R., Mayhaus, M., Lannefelt, L., Hakonarson, H., Pichler, S., Carrasquillo, M.M., Ingelsson, M., Beekly, D., Alvarez, V., Zou, F., Valladares, O., Younkin, S.G., Coto, E., Hamilton-Nelson, K.L., Gu, W., Razquin, C., Pastor, P., Mateo, I., Owen, M.J., Faber, K.M., Jonsson, P.V., Combarros, O., O’Donovan, M.C., Cantwell, L.B., Soininen, H., Blacker, D., Mead, S., Mosley Jr., T.H., Bennett, D.A., Harris, T.B., Fratiglioni, L., Holmes, C., de Bruijn, R.F., Passmore, P., Montine, T.J., Bettens, K., Rotter, J.I., Brice, A., Morgan, K., Foroud, T.M., Kukull, W.A., Hannequin, D., Powell, J.F., Nalls, M.A., Ritchie, K., Lunetta, K.L., Kauwe, J.S., Boerwinkle, E., Riemenschneider, M., Boada, M., Hiltuenen, M., Martin, E.R., Schmidt, R., Rujescu, D., Wang, L.S., Dartigues, J.F., Mayeux, R., Tzourio, C., Hofman, A., Nothen, M.M., Graff, C., Psaty, B.M., Jones, L., Haines, J.L., Holmans, P.A., Lathrop, M., Pericak-Vance, M.A., Launer, L.J., Farrer, L.A., van Duijn, C.M., Van Broeckhoven, C., Moskvina, V., Seshadri, S., Williams, J., Schellenberg, G.D. and Amouyel, P. (2013) Meta-Analysis of 74,046 Individuals Identifies 11 New Susceptibility Loci for Alzheimer’s Disease. Nature Genetics, 45, 1452-1458.
- 9. International Multiple Sclerosis Genetics Consortium (2013) Network-Based Multiple Sclerosis Pathway Analysis with GWAS Data from 15,000 Cases and 30,000 Controls. American Journal of Human Genetics, 92, 854-865. https://doi.org/10.1016/j.ajhg.2013.04.019
- 10. Goel, R., Harsha, H.C., Pandey, A. and Prasad, T.S. (2012) Human Protein Reference Database and Human Proteinpedia as Resources for Phosphoproteome Analysis. Molecular BioSystems, 8, 453-463. https://doi.org/10.1039/C1MB05340J
- 11. Huang, D.W., Sherman, B.T., Tan, Q., Kir, J., Liu, D., Bryant, D., Guo, Y., Stephens, R., Baseler, M.W., Lane, H.C. and Lempicki, R.A. (2007) DAVID Bioinformatics Resources: Expanded Annotation Database and Novel Algorithms to Better Extract Biology from Large Gene Lists. Nucleic Acids Research, 35, W169-W175. https://doi.org/10.1093/nar/gkm415
- 12. Kanehisa, M. and Goto, S. (2000) KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Research, 28, 27-30. https://doi.org/10.1093/nar/28.1.27
- 13. Heun, R., Kolsch, H. and Jessen, F. (2006) Risk Factors and Early Signs of Alzheimer’s Disease in a Family Study Sample. Risk of AD. European Archives of Psychiatry and Clinical Neuroscience, 256, 28-36. https://doi.org/10.1007/s00406-005-0596-4
- 14. Zick, C.D., Smith, K.R. and Mayer, R.N. (2016) Planning Ahead or Living a Day at a Time? A Family History of AD and Retirement Planning. American Journal of Alzheimer's Disease and Other Dementias, 31, 516-523. https://doi.org/10.1177/1533317516653821
- 15. Takashima, A. (2015) Toxic Tau Aggregation in AD. Advances in Experimental Medicine and Biology, 822, 3-9. https://doi.org/10.1007/978-3-319-08927-0_2
- 16. Carrera, I., Etcheverria, I., Li, Y., Fernandez-Novoa, L., Lombardi, V., Vigo, C., Palacios, H.H., Benberin, V.V., Cacabelos, R. and Aliev, G. (2013) Immunocytochemical Characterization of Alzheimer Disease Hallmarks in APP/PS1 Transgenic Mice Treated with a New Anti-Amyloid-Beta Vaccine. BioMed Research International, 2013, Article ID: 709145. https://doi.org/10.1155/2013/709145
- 17. Frankish, H. (2005) Unravelling the Mechanism of Amyloid-Beta Toxicity in AD. The Lancet Neurology, 4, 526. https://doi.org/10.1016/S1474-4422(05)70153-8
- 18. Stefanova, E., Blennow, K., Almkvist, O., Hellstrom-Lindahl, E. and Nordberg, A. (2003) Cerebral Glucose Metabolism, Cerebrospinal Fluid-Beta-Amyloid1-42 (CSF-Abeta42), Tau and Apolipoprotein E Genotype in Long-Term Rivastigmine and Tacrine Treated Alzheimer Disease (AD) Patients. Neuroscience Letters, 338, 159-163. https://doi.org/10.1016/S0304-3940(02)01384-8
- 19. Pocernich, C.B. and Butterfield, D.A. (2003) Acrolein Inhibits NADH-Linked Mitochondrial Enzyme Activity: Implications for Alzheimer’s Disease. Neurotoxicity Research, 5, 515-520. https://doi.org/10.1007/BF03033161
- 20. Manczak, M., Park, B.S., Jung, Y. and Reddy, P.H. (2004) Differential Expression of Oxidative Phosphorylation Genes in Patients with Alzheimer’s Disease: Implications for Early Mitochondrial Dysfunction and Oxidative Damage. NeuroMolecular Medicine, 5, 147-162. https://doi.org/10.1385/NMM:5:2:147
- 21. Grazina, M., Silva, F., Santana, I., Pratas, J., Santiago, B., Oliveira, M., Carreira, I., Cunha, L. and Oliveira, C. (2005) Mitochondrial DNA Variants in a Portuguese Population of Patients with Alzheimer’s Disease. European Neurology, 53, 121-124. https://doi.org/10.1159/000085555
- 22. Breier, A., Bohacova, V. and Docolomansky, P. (2006) Inhibition of (Na(+)/K(+))-ATPase by Cibacron Blue 3G-A and Its Analogues. General Physiology and Biophysics, 25, 439-453.
- 23. Chandrasekaran, K., Giordano, T., Brady, D.R., Stoll, J., Martin, L.J. and Rapoport, S.I. (1994) Impairment in Mitochondrial Cytochrome Oxidase Gene Expression in Alzheimer Disease. Brain Research. Molecular Brain Research, 24, 336-340. https://doi.org/10.1016/0169-328X(94)90147-3
- 24. Pappolla, M.A., Omar, R.A., Chyan, Y.J., Ghiso, J., Hsiao, K., Bozner, P., Perry, G., Smith, M.A. and Cruz-Sanchez, F. (2001) Induction of NADPH Cytochrome P450 Reductase by the Alzheimer Beta-Protein. Amyloid as a “Foreign Body”. Journal of Neurochemistry, 78, 121-128. https://doi.org/10.1046/j.1471-4159.2001.00379.x
- 25. Schagger, H. and Ohm, T.G. (1995) Human Diseases with Defects in Oxidative Phosphorylation. 2. F1F0 ATP-Synthase Defects in Alzheimer Disease Revealed by Blue Native Polyacrylamide Gel Electrophoresis. European Journal of Biochemistry, 227, 916-921. https://doi.org/10.1111/j.1432-1033.1995.0916p.x
- 26. Wood, H. (2014) Alzheimer Disease: Alzheimer Disease Risk Factor CALM Modulates Tau Turnover. Nature Reviews Neurology, 10, 610. https://doi.org/10.1038/nrneurol.2014.188
