Computer Science and Application
Vol.
11
No.
03
(
2021
), Article ID:
40892
,
13
pages
10.12677/CSA.2021.113048
多功能酶的分类技术与应用
毕鹏丽
云南大学信息学院,云南 昆明
收稿日期:2021年2月8日;录用日期:2021年3月3日;发布日期:2021年3月12日

摘要
酶是由活细胞产生的、对其底物具有高度特异性和高度催化效能的蛋白质或RNA,具有多种催化功能的酶被称为多功能酶。细胞是高度精细的复杂有机网络,多功能酶是常见的重要代谢反应的参与者,参与多个细胞代谢网络。在数据挖掘和机器学习领域,对酶的研究可以看作是一项预测任务。本文从机器学习的角度对关于多功能酶的研究作了一个深入的回顾。从方法和应用的角度,讨论的建模方法包括数据预处理、分类算法和模型评估等技术。对于应用方面,对现有的多功能酶应用领域提供了一个全面的分类,然后对各类别的应用进行了详细说明。最后,结合经验和判断,总结了一些建议,为多功能酶领域的进一步研究提供了方向。
关键词
生物信息学,多功能酶,多标签,机器学习
Classification Technology and Application of Multifunctional Enzymes
Pengli Bi
School of Information Science and Engineering, Yunnan University, Kunming Yunnan
Received: Feb. 8th, 2021; accepted: Mar. 3rd, 2021; published: Mar. 12th, 2021

ABSTRACT
Enzymes are proteins or RNAs produced by living cells, which are highly specific and highly catalytic for their substrates. Enzymes with multiple catalytic functions are called multifunctional enzymes. Cells are highly sophisticated and complex organic networks, and multifunctional enzymes are common participants in important metabolic reactions and participate in multiple cellular metabolic networks. In the field of data mining and machine learning, the research of enzymes can be regarded as a prediction task. The article provides an in-depth review of the research on enzymes from the perspective of machine learning. From the perspective of methods and applications, the modeling methods discussed include data preprocessing, classification algorithms, and model evaluation. For application, a comprehensive classification is provided for the existing multifunctional enzyme application fields, and then the application of each category is described in detail. Finally, combined with experience and judgment, some suggestions in the paper are summarized, which provides a direction for further research in the field of multifunctional enzymes.
Keywords:Bioinformatics, Multifunctional Enzyme, Multi-Label, Machine Learning

Copyright © 2021 by author(s) and Hans Publishers Inc.
This work is licensed under the Creative Commons Attribution International License (CC BY 4.0).
http://creativecommons.org/licenses/by/4.0/


1. 引言
1999年Jeffery将一类同时具有两种或两种以上功能的蛋白质称为兼职蛋白质 [1],还对兼职蛋白质的概念进行了严格的限定。经过研究表明,功能明确的兼职蛋白质大部分是酶蛋白,即除了已经具备的催化功能外还具有其他的生物学功能,这些功能涵盖了两种情况:一是产生催化混杂的现象 [2],二是与调节作用有关的功能。这类具有多种功能的酶被称为多功能酶。在数据挖掘领域,关于多功能酶的研究是一个预测问题,通常是一个数据分类问题。虽然数据挖掘方法已被广泛应用于建立分类模型来指导预测任务,但对多功能酶这类多标签的分类模型构成了重大的挑战。多标签分类与传统的分类问题相比较,主要难点有以下两个:
1) 类标签的数量不确定,有的样本可能只有一个类标签,而有的样本的类标签可能高达几十个;
2) 类标签之间存在依赖,怎样解决类标签之间的依赖性也是一个难点。
为了解决多功能酶分类问题,在过去人们已经开发了机器学习方法,其中大部分是基于样本技术和集成方法等 [3]。尽管已经发表了一些与多功能酶分类有关的研究 [4] [5] [6],但大都集中于技术方面,而忽略了应用方面的探索,关于多功能酶这个大类的应用研究综述少之又少。对于生物学等领域的研究人员来说,使用先进的学习技术和成熟有效的方法解决多功能酶分类预测的问题可能比使用复杂的算法更为重要。
本文目的是提供一个关于多功能酶研究的全面概述,包括方法和应用两方面。在方法层面,介绍了研究多功能酶的常用方法,包括预处理、分类和评价。在应用方面,回顾了多功能酶在不同研究领域的应用。最后我们从方法和应用两方面讨论了未来对于多功能酶研究方向的思考,并在第5节进行了全文总结。
2. 研究方法和初步统计
2.1. 研究方法
本研究是基于Govindan和Jepsen [7] 的研究方法,采用两阶段检索程序,收集了近年发表的相关论文。在初始阶段,我们使用图书馆数据库进行文献检索和收集,并按照Fahimnia等人(2015) [8] 概述的搜索过程设计搜索词。图1给出了本研究的搜索词。第一阶段的搜索限于多功能酶,我们从多个数据库中搜索带有“多功能酶”字段的文献。第二阶段的搜索分两部分,包含方法和应用。在方法方面,使用了数据挖掘方法中的关键词,如:机器学习、深度学习、多标签学习等,在实际应用方面,使用了应用领域的关键词,如:疾病、癌症、预后、病毒、表达作用。
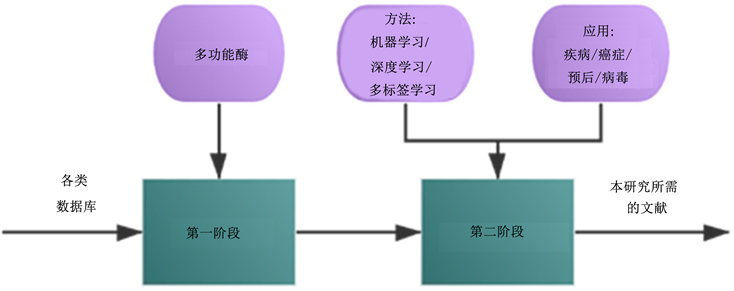
Figure 1. Two-stage keywords tree structure
图1. 两阶段关键树结构
2.2. 初步统计
在手工审阅每篇论文后,共发现89篇与本研究相关的论文。在本节中,我们将介绍多功能酶研究的初步统计数据。图2显示了2010~2020年出版数量的变化趋势。这种趋势表明,多功能酶研究仍然是一个有价值的研究课题。

Figure 2. Publication trend of multifunctional enzyme research
图2. 多功能酶研究发表趋势
初步统计,共有60多种期刊和会议论文集发表了关于多功能酶研究的文章。这些期刊包含了计算机科学、肿瘤学、微生物学、工程和生物技术等领域。根据每一种期刊的贡献度,排名前13的期刊或者会议的统计结果如图3所示。这也说明多功能酶研究既包括计算机的学习技术,也包括从自然科学到工程科学等广泛领域的实际应用。

Figure 3. Top 13 journals/conferences on which most multifunctional enzyme research
图3. 多功能酶研究论文发表最多的13种期刊/会议
我们还收集了本次研究中涉及的所有论文的题目,并生成单词云,以捕捉研究中最多的多功能酶研究主题。为了构建图4所示的词云,我们首先去除英语中最常用的停止词,如“the”和“and”,然后对每个词进行归类分析。由于我们的目标是在这些多功能酶研究的论文中发现详细的主题,所以一些一般性和经常出现的词汇也被删除。图4显示了多功能酶研究中的一些具体技术:机器学习方法(“支持向量机”、“多标签学习”、“集成”等),并且一些具体的应用领域也显示在单词云中,如“肿瘤”、“病毒”、“癌症”、“预后”、“畜牧”等,这意味着已发表论文的主要类别属于医疗诊断、术后治愈、养殖工程等领域。

Figure 4. Word cloud of the title words from the collected paper
图4. 收集论文中标题词的词云
3. 解决多功能酶分类问题的基本策略
3.1. 预处理
所有酶序列均来自于http://enzyme.expasy.org/网站上的酶命名数据库。为了获得更好的输入数据来构建一个高更新基准数据集,开发一个预测器来识别酶的类别,通常在建立学习模型之前进行预处理。一般考虑以下步骤:
步骤1:仅收集关键词为“多功能酶”的序列;
步骤2:删除标注“fragment”的序列;
步骤3:根据实验需要,去除长度小于N的氨基酸残基的序列,因为这些序列可能属于片段;
步骤4:为了减少冗余性和同源性的影响,我们通常使用CD-HIT程序排除这些酶与同一亚群中任何其他酶在成对序列相似度超过m%的酶(m根据实验需求设定,最小为40)。
3.2. 多功能酶样本的表示
为了建立一种基于序列信息预测多功能酶的有效预测器,其中最重要的一步就是特征选择与特征提取,即用有效的数学表达式构建序列,并且该序列能够真实地反映与待识别目标的内在相关性。以下为多功能酶研究中常见的几种特征提取方法。
3.2.1. 氨基酸组成(AAC)
蛋白质序列由20种常见的氨基酸组成,氨基酸组成的特征表达就是计算各种氨基酸在待测蛋白质序列中出现的频率 [9]。虽然用AAC可以预测很多蛋白质属性 [10] [11],但是存在一个致命缺点,即如果使用AAC作为唯一的特征提取方法来提取蛋白质信息,则其所有的序列顺序和序列长度信息将丢失。因此为了避免这样的情况发生,Chou [12] 提出了伪氨基酸组成(PseAAC)来代替氨基酸组成(AAC)。
3.2.2. 伪氨基酸组成(PseAAC)
伪氨基酸组成最早是由Chou在2001年提出来的,随后PseAAC的思想广泛应用于蛋白质组学、生物信息学和系统生物学 [13],比如预测蛋白质亚细胞定位 [14],预测DNA结合蛋白质 [15],氨基酸的分类 [16],预测酶家族分类 [17],预测蛋白质四级结构属性 [18],鉴别外膜蛋白 [19]。目前,PseAAC被用来处理DNA领域的问题,例如识别核小体 [20] 和预测重组点 [21]。
3.2.3. SAAC (Split Amino Acid Composition)
根据SAAC方法,一条蛋白质序列分为不同的部分并各自计算每个部分的组成。鉴于 [22] 此,一条蛋白质序列被分为三个部分 [23]:N端、中间段、C段。因为在对多功能酶数据进行筛选时,把氨基酸残基少于50的蛋白质序列已经去除了,所以在SAAC中,N端和C端分别包括25个氨基酸,其余的氨基酸在中间段中,然后分别计算这三段蛋白质序列中各氨基酸出现的频率。一般来说,特征选择的目标是从整个特征空间中选择k个特征的子集,使分类器达到最优性能。处理数据的另一种方法是特征提取。特征提取与降维有关,降维是将数据转换为低维空间。然而,应该注意的是,特征选择技术不同于特征提取技术。特征提取使用函数映射从原始特征创建新特征,而特征选择返回原始特征的子集 [24]。对图像、文本和语音等非结构化数据,特征提取方法的应用越来越多。表1显示了使用特征选择或特征提取的文章。我们发现,特征选择和特征提取经常用于解决现实世界的问题,如疾病诊断、癌症治疗等。
Table 1. Summary of articles employing feature selection or extraction methods
表1. 使用特征选择或提取方法的文章总结
3.3. 分类算法
针对多功能酶研究的多标签学习系统,一个良好的多标签分类算法对预测效果有着至关重要的作用。多标签数据学习方法依据问题解决的角度,主要分为两种。
(一) 问题转换法。基于问题转换的方法中有些考虑类标之间的依赖性,有些不考虑。最常见的不考虑类标之间依赖性的方法是将多标签问题进行分解,将其转换成n个二元分类问题(n是类别个数),将多标签中的每一个标签看作是单标签,然后对每一个标签实施常见的分类算法 [30],表2列举了问题转换方法中的常用方式及其优缺点;
Table 2. Common methods in problem conversion methods
表2. 问题转换方法中的常用方式
(二) 算法适应法。基于算法适应的方法是针对某一特定算法进行扩展 [31],进而改进算法使得能够处理多标签数据。在传统机器学习模型中常见的多标签分类模型如图5所示。表3列出了相关算法的使用情况。
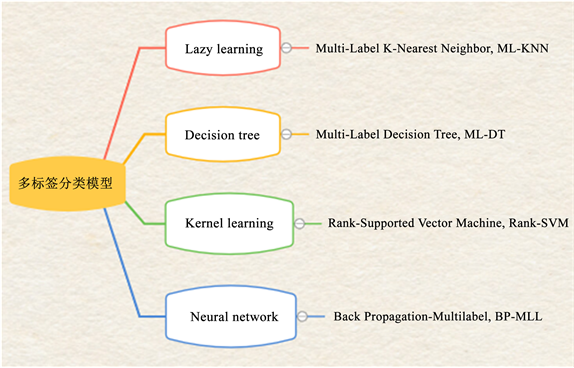
Figure 5. Common multi-label classification models
图5. 常见的多标签分类模型
Table 3. Representative article on classification algorithm
表3. 分类算法的代表文章
3.4. 评价指标
模型选择和模型评价是机器学习中的两个关键过程。因此,性能度量是评价分类器有效性和指导分类器学习的关键指标。多标签学习系统的性能评价不同于传统的单标签学习系统。在单标签系统中常用的评价指标包括:精确率、正确率、召回率和F-Score等 [39],但对于多标签学习系统这些评价指标要
复杂得多,例如: ,,。
4. 多功能酶的应用
本节的目的是向来自不同领域的研究人员展示多功能酶问题如何应用于研究领域。由于方法技术已经在第三节中介绍过了,我们在本节中尽量省略方法细节,以避免重复,经过整理,我们发现49篇从疾病到工程应用导向的文章是。我们将49篇文章分为4个应用领域。在第3.2中,每个类别都将详细描述并给出一些例子。
4.1. 应用领域的分类
我们将多功能酶研究的应用领域分为4大类:疾病、癌症和肿瘤、病毒、其他。其他类别包括生物燃料、生物合成、畜牧等。表4显示了分类以及应用文章的频次分布。
Table 4. Application domain categories
表4. 应用领域分类
4.2. 各个领域的详细介绍
(一) 多功能酶在疾病领域的研究热点主要包括:预后治疗和控制影响某种疾病的激素,如通过控制胰岛素对糖尿病人进行治疗;
(二) 经过审查,发现有14篇文章与肿瘤癌症有关,主要的应用包含:在各类癌症中表达作用以及临床意义、靶点治疗、在肿瘤细胞中的增殖、凋亡作用。我们单独把癌症肿瘤从疾病这一应用领域分出来,是因为多功能酶在疾病中的应用主要与普通疾病有关,如:白血病、糖尿病等。而在癌症方面的应用相对来说更多,如:肝癌、乳腺癌、前列腺癌、胃癌、宫颈癌、膀胱癌、卵巢癌、食管癌子宫内膜癌等,多功能酶在这些癌症治疗过程中都发挥着靶点治疗的重要作用;
(三) 在病毒这一领域的应用,近年来主要讨论了多功能酶与登革病毒的关系;
(四) 其他应用领域主要包括:生物工程(生物合成、生物燃料等)、畜牧、纺织等,尤其在水产养殖方面有很大的实用价值,不仅提高产量还提高质量。相关研究问题及参考文献见表5。
Table 5. Applications in various fields
表5. 各个领域的应用
5. 总结与展望
在本文中,我们对多功能酶分类技术及其应用进行了全面的概述,在此过程中我们从方法和应用的角度出发,收集了89篇与多功能酶研究相关的论文。根据调查结果,我们提出了多功能酶研究未来可能的研究方向。
(一) 方法方面:可以尝试其他的特征选择方法获得我们所需要的实验数据,然后经过特征融合的方法将特征进行融合使我们的实验数据能够更好的表达它的意义;此外作为一种提高弱学习者分类性能的好方法,基于集成的算法目前已被广泛用于解决一些多分类学习任务。Wang and Yao [89] 认为集成模型的性能取决于单个分类器的准确性和所有分类器之间的多样性,未来可以尝试将多标签学习算法中的分类器进行集成,以获得更好的性能。往后可以将重心放在多标签学习两大难点上,开发新的算法去解决标签依赖性以及类标不平衡的问题。
(二) 应用方面:回顾第3节的应用分布,只有2篇文献讨论了多功能酶在病毒研究领域方面的应用,由于新药研发已经进入到了以生物靶标为核心的时代,特别是以计算机模拟结构来设计药物的出现使得药物设计理念得到了进一步的完善,未来可以在这个方面做更多的工作;另一个有价值的研究方向是在其他应用领域中,多功能酶的应用较广,从生物合成到畜牧水产都有涉猎,尤其近年来多功能酶在畜牧、水产、养蜂方面有很大的应用成果。现在人们更加注重健康与营养,怎样提高产品的产量与质量也是一个需要继续攻克的难题。
基金项目
国家自然科学基金(62062067)。
文章引用
毕鹏丽. 多功能酶的分类技术与应用
Classification Technology and Application of Multifunctional Enzymes[J]. 计算机科学与应用, 2021, 11(03): 476-488. https://doi.org/10.12677/CSA.2021.113048
参考文献
- 1. Jeffery, C.J. (2003) Moonlighting Proteins: Old Proteins Learning New Tricks. Trends in Genetics, 19, 415-417. https://doi.org/10.1016/S0168-9525(03)00167-7
- 2. Moore, B. (2004) Bifunctional and Moonlighting Enzymes: Lighting the Way to Regulatory Control. Trends in Plant Science, 9, 221-228. https://doi.org/10.1016/j.tplants.2004.03.005
- 3. Zou, H.L. and Xiao, X. (2016) Classifying Multifunctional En-zymes by Incorporating Three Different Models into Chou’s General Pseudo Amino Acid Composition. The Journal of Membrane Biology, 249, 551-557. https://doi.org/10.1007/s00232-016-9904-3
- 4. 黄炜娟. 多功能酶的预测及结构功能模式分析[D]: [硕士学位论文]. 厦门: 厦门大学, 2009.
- 5. 王跃龙, 敬闰宇, 李梦龙, 华勇攀. 基于多标签机器学习和序列描述符对多功能酶进行功能预测[C]//中国化学会. 中国化学会第29届学术年会摘要集——第19分会: 化学信息学与化学计量学. 中国化学会, 2014: 1.
- 6. 刘干. 多功能酶分类预测中的特征表达与融合算法研究[D]: [硕士学位论文]. 云南: 云南大学, 2019.
- 7. Govindan, K. and Jepsen, M.B. (2016) ELECTRE: A Comprehensive Literature Review on Methodologies and Applications. European Journal of Operational Research, 250, 1-29. https://doi.org/10.1016/j.ejor.2015.07.019
- 8. Fahimnia, B., Tang, C.S., Davarzani, H. and Sarkis, J. (2015) Quantitative Models for Managing Supply Chain Risks: A Review. European Journal of Operational Research, 247, 1-15. https://doi.org/10.1016/j.ejor.2015.04.034
- 9. Liu, S., Wang, S.F. and Ding, H. (2015) Protein Sub-Nuclear Lo-cation by Fusing AAC and PSSM Features Based on Sequence Information. 2015 IEEE 5th International Conference on Electronics Information and Emergency Communication, Beijing, 14-16 May 2015, 236-239. https://doi.org/10.1109/ICEIEC.2015.7284529
- 10. Zhou, G.P. and Assa-Munt, N. (2001) Some Insights into Protein Structural Class Prediction. Proteins: Structure, Function, and Bioinformatics, 44, 57-59. https://doi.org/10.1002/prot.1071
- 11. Zhou, G.-P. and Doctor, K. (2003) Subcellular Location Prediction of Apoptosis Proteins. Proteins: Structure, Function, and Bioinformatics, 50, 44-48. https://doi.org/10.1002/prot.10251
- 12. Chou, K.-C. (2001) Prediction of Protein Cellular Attributes Using Pseu-do-Amino Acid Composition. Proteins: Structure, Function, and Bioinformatics, 44, 60. https://doi.org/10.1002/prot.1072
- 13. Chou, K.-C. (2009) Pseudo Amino Acid Composition and Its Applications in Bioinformatics, Proteomics and System Biology. Current Proteomics, 6, 262-274. https://doi.org/10.2174/157016409789973707
- 14. Zhang, S.-W., Zhang, Y.-L., Yang, H.-F., Zhao, C.-H. and Pan, Q. (2008) Using the Concept of Chou’s Pseudo Amino Acid Composition to Predict Protein Subcellular Localiza-tion: An Approach by Incorporating Evolutionary Information and Von Neumann Entropies. Amino Acids, 34, 565-572. https://doi.org/10.1007/s00726-007-0010-9
- 15. Lin, W.-Z., Fang, J.-A., Xiao, X. and Chou, K.-C. (2011) iD-NA-Prot: Identification of DNA Binding Proteins Using Random Forest with Grey Model. PLoS ONE, 6, e24756. https://doi.org/10.1371/journal.pone.0024756
- 16. Georgiou, D.N., Karakasidis, T.E., Nieto, J.J. and Torres, A. (2009) Use of Fuzzy Clustering Technique and Matrices to Classify Amino Acids and Its Impact to Chou’s Pseudo Amino Acid Composition. Journal of Theoretical Biology, 257, 17-26. https://doi.org/10.1016/j.jtbi.2008.11.003
- 17. Zhou, X.-B., Chen, C., Li, Z.-C. and Zou, X.-Y. (2007) Using Chou’s Amphiphilic Pseudo-Amino Acid Composition and Support Vector Machine for Prediction of Enzyme Subfami-ly Classes. Journal of Theoretical Biology, 248, 546-551. https://doi.org/10.1016/j.jtbi.2007.06.001
- 18. Sun, X.-Y., Shi, S.-P., Qiu, J.-D., Suo, S.-B., Huang, S.-Y. and Liang, R.-P. (2012) Identifying Protein Quaternary Structural Attributes by Incorporating Physicochemical Properties into the General form of Chou’s PseAAC via Discrete Wavelet Transform. Molecular BioSystems, 8, 3178-3184. https://doi.org/10.1039/c2mb25280e
- 19. Hayat, M. and Khan, A. (2012) Discriminating Outer Membrane Proteins with Fuzzy K-Nearest Neighbor Algorithms Based on the General Form of Chou’s PseAAC. Protein & Peptide Letters, 19, 411-421. https://doi.org/10.2174/092986612799789387
- 20. Chen, W., Lin, H., Feng, P.-M., Ding, C., Zuo, Y.-C. and Chou, K.-C. (2017) iNuc-PhysChem: A Sequence-Based Predictor for Identifying Nucleosomes via Physicochemical Properties. PLoS ONE, 7, e47843. https://doi.org/10.1371/journal.pone.0047843
- 21. Chen, W., Feng, P.-M., Lin, H. and Chou, K.-C. (2013) iR-Spot-PseDNC: Identify Recombination Spots with Pseudo Dinucleotide Composition. Nucleic Acids Research, 41, e68. https://doi.org/10.1093/nar/gks1450
- 22. Chou, K.-C. and Shen, H.-B. (2006) Predicting Eukaryotic Protein Sub-cellular Location by Fusing Optimized Evidence-Theoretic K-Nearest Neighbor Classifiers. Journal of Proteome Re-search, 5, 1888-1897.
- 23. Hayat, M., Khan, A. and Yeasin, M. (2012) Prediction of Membrane Proteins Using Split Amino Acid and Ensemble Classification. Amino Acids, 42, 2447-2460. https://doi.org/10.1007/s00726-011-1053-5
- 24. Motoda, H. and Liu, H. (2002) Feature Selection, Extraction and Construction. Communication of IICM (Institute of Information and Computing Machinery, Taiwan), 5, 67-72.
- 25. Blagus, R. and Lusa, L. (2010) Class Prediction for High-Dimensional Class-Imbalanced Data. BMC Bioin-formatics, 11, Article No. 523. https://doi.org/10.1186/1471-2105-11-523
- 26. Yang, J.S., Zhou, J.R., Zhu, Z.X., Ma, X.L. and Ji, Z. (2016) Iterative Ensemble Feature Selection for Multiclass Classification of Imbalanced Microarray Data. Journal of Biological Research-Thessaloniki, 23, Article No. 13. https://doi.org/10.1186/s40709-016-0045-8
- 27. Hai, P.-T., Gerardo, C.-M., Teresa, G., Bermejo, M., Gonzá-lez-Álvarez, I., Nguyen-Hai, N., et al. (2016) Exploring Different Strategies for Imbalanced ADME Data Problem: Case Study on Caco-2 Permeability Modeling. Molecular Diversity, 20, 93-109. https://doi.org/10.1007/s11030-015-9649-4
- 28. Dubey, R., Zhou, J., Wang, Y., Thompson, P.M., Ye, J. and the Alzheimer’s Disease Neuroimaging Initiative (2014) Analysis of Sampling Techniques for Imbalanced Data: An n = 648 ADNI Study. NeuroImage, 87, 220-241. https://doi.org/10.1016/j.neuroimage.2013.10.005
- 29. Zhang, N. (2016) Cost-Sensitive Spectral Clustering for Photo-Thermal Infrared Imaging Data. 2016 Sixth International Conference on Information Science and Technology, Da-lian, 6-8 May 2016, 358-361. https://doi.org/10.1109/ICIST.2016.7483438
- 30. 张泉灵. 多标记学习研究综述[J]. 计算机工程与应用, 2015, 51(17): 20-27.
- 31. 冯雪东. 多标签分类问题综述[J]. 信息系统工程, 2016(3): 137.
- 32. Zhang, M.-L. and Zhou, Z.-H. (2007) ML-KNN: A Lazy Learning Approach to Multi-Label Learning. Pattern Recognition, 40, 2038-2048. https://doi.org/10.1016/j.patcog.2006.12.019
- 33. 孙鑫亮, 杨涛, 章颖, 董海艳, 胡孔法, 谢佳东, 史话跃. 基于ML-KNN算法的冠心病辨证模型研究[J]. 山东中医药大学学报, 2019, 43(5): 438-442.
- 34. Jiang, M., et al. (2019) A Classification Algorithm Based on Weighted ML-kNN for Multi-Label Data. International Journal of Internet Manufacturing and Services, 6, Article No. 4. https://doi.org/10.1504/IJIMS.2019.103861
- 35. Panesar, S.S., D’Souza, R.N., Yeh, F.-C. and Fernandez-Miranda, J.C. (2019) Machine Learning Versus Logistic Regression Methods for 2-Year Mortality Prognostication in a Small, Heterogeneous Glioma Database. World Neurosurgery: X, 2, Article ID: 100012. https://doi.org/10.1016/j.wnsx.2019.100012
- 36. Albaradei, S., Magana-Mora, A., Thafar, M., Uludag, M., Bajic, V.B., Gojobori, T., et al. (2020) Splice2Deep: An Ensemble of Deep Convolutional Neural Networks for Im-proved Splice Site Prediction in Genomic DNA. Gene: X, 5, Article ID: 100035. https://doi.org/10.1016/j.gene.2020.100035
- 37. 辛基梁, 李绍滋, 张佳, 雷黄伟, 李灿东. 中医健康状态辨识方法的探索[J]. 中华中医药杂志, 2019, 34(7): 3151-3153.
- 38. Zhang, K.L., Ma, H.C., Zhao, Y.S., Zan, H.Y. and Zhuang, L. (2018) The Comparative Experimental Study of Multilabel Classification for Diagnosis Assistant Based on Chinese Obstetric EMRs. Journal of Healthcare Engineering, 2018, Article ID: 7273451. https://doi.org/10.1155/2018/7273451
- 39. Sebastiani, F. (2018) Machine Learning in Automated Text Categoriza-tion. ACM Computing Surveys, 34, 1-47. https://doi.org/10.1145/505282.505283
- 40. Maryam, G.H., Soheila, R., Somayeh, R.B., Safavi, A. and Ghodousi, E.S. (2020) Publisher Correction: Fatty Acid Synthase, a Novel Poor Prognostic Factor for Acute Lymphoblastic Leu-kemia which Can Be Targeted by Ginger Extract. Scientific Reports, 10, Article No. 20952. https://doi.org/10.1038/s41598-020-78089-5
- 41. 占敏霞, 巫冠中. 脂肪酸合成酶与疾病[J]. 亚太传统医药, 2012, 8(9): 210-212.
- 42. 王冰, 高政南. 脂肪酸合成酶在糖尿病小鼠肾脏的表达及胰岛素的调节作用[C]// 中华医学会. 中华医学会糖尿病学分会第十六次全国学术会议论文集2012年卷. 中华医学会, 2012: 2.
- 43. 王冰, 程丽静, 刘明川, 高政南. 脂肪酸合成酶和碳水化合物反应元件结合蛋白在1型糖尿病小鼠肾脏的表达及胰岛素对其的调节作用[J]. 中国糖尿病杂志, 2016, 24(1): 64-68.
- 44. Hopperton, K.E., Duncan, R.E., Bazinet, R.P. and Archer, M.C. (2014) Fatty Acid Synthase Plays a Role in Cancer Metabolism beyond Providing Fatty Acids for Phos-pholipid Synthesis or Sustaining Elevations in Glycolytic Activity. Experimental Cell Research, 320, 302-310. https://doi.org/10.1016/j.yexcr.2013.10.016
- 45. Hao, Q.W., Li, T., Zhang, X., Gao, P., Qiao, P.Y., Li, S., et al. (2014) Expression and Roles of Fatty Acid Synthase in Hepatocellular Carcinoma. Oncology Reports, 32, 2471-2476. https://doi.org/10.3892/or.2014.3484
- 46. Allina, D.O., Andreeva, Y.Y., Zavalishina, L.E., Moskvina, L.V. and Frank, G.A. (2017) Fatty Acid Synthase in the Diagnosis of Prostate Neoplasms. Arkhivpatologii, 79, 10-14. https://doi.org/10.17116/patol201779210-14
- 47. Yuan, Y.J., Yang, X.Q., Li, Y., Liu, Q., Wu, F., Qu, H.Y., et al. (2020) Expression and Prognostic Significance of Fatty Acid Synthase in Clear Cell Renal Cell Carcinoma. Patholo-gy-Research and Practice, 216, Article ID: 153227. https://doi.org/10.1016/j.prp.2020.153227
- 48. Song, L.-R., Li, D., Weng, J.-C., Li, C.-B., Wang, L., Wu, Z., et al. (2020) MicroRNA-195 Functions as a Tumor Suppressor by Directly Targeting Fatty Acid Synthase in Malignant Men-ingioma. World Neurosurgery, 136, e355- e364. https://doi.org/10.1016/j.wneu.2019.12.182
- 49. Hirai, H., Tada, Y., Nakaguro, M., Kawakita, D., Sato, Y., Shimura, T., et al. (2020) The Clinicopathological Significance of the Adipophilin and Fatty Acid Synthase Expression in Salivary Duct Carcinoma. Virchows Archiv, 477, 291-299. https://doi.org/10.1007/s00428-020-02777-w
- 50. Jia, J.Y., et al. (2020) Pivotal Role of Fatty Acid Synthase in c-MYC Driven Hepatocarcinogenesis. International Journal of Molecular Sciences, 21, 8467. https://doi.org/10.3390/ijms21228467
- 51. Menendez, J.A., Mehmi, I., Papadimitropoulou, A., Vander Steen, T., Cuyàs, E., Verdura, S., et al. (2020) Fatty Acid Synthase Is a Key Enabler for Endocrine Resistance in Heregulin-Overexpressing Luminal B-Like Breast Cancer. International Journal of Molecular Sciences, 21, 7661. https://doi.org/10.3390/ijms21207661
- 52. Jia, J.Y., Che, L., Cigliano, A., Wang, X., Peitta, G, Tao, J.Y., et al. (2020) Pivotal Role of Fatty Acid Synthase in c-MYC Driven Hepatocarcinogenesis. International Journal of Molecular Sciences, 21, 8467.
- 53. Rahman, M.T., Nakayama, K., Ishikawa, M., Rahman, M., Katagiri, H, Katagiri, A., et al. (2013) Fatty Acid Synthase Is a Potential Therapeutic Target in Estrogen Receptor-/Progesterone Re-ceptor-Positive Endometrioid Endometrial Cancer. Oncology, 84, 166-173. https://doi.org/10.1159/000342967
- 54. Menendez, J.A. and Lupu, R. (2017) Fatty Acid Synthase (FASN) as a Therapeutic Target in Breast Cancer. Expert Opinion on Therapeutic Targets, 21, 1001-1016. https://doi.org/10.1080/14728222.2017.1381087
- 55. Yoshii, Y., Furukawa, T., Oyama, N., Hasegawa, Y., Ki-yono, Y., Nishii, R., et al. (2017) Fatty Acid Synthase Is a Key Target in Multiple Essential Tumor Functions of Prostate Cancer: Uptake of Radiolabeled Acetate as a Predictor of the Targeted Therapy Outcome. PLoS ONE, 8, e64570. https://doi.org/10.1371/journal.pone.0064570
- 56. Walz, J.Z., Saha, J., Arora, A., Khammanivong, A., O’Sullivan, M.G. and Dickerson, E.B. (2018) Fatty Acid Synthase as a Potential Therapeutic Target in Feline Oral Squamous Cell Carcinoma. Veterinary and Comparative Oncology, 16, E99-E108. https://doi.org/10.1111/vco.12341
- 57. Li, C., Panagiotis, P., Antonio, C., Pilo, M.G., Chen, X. and Calvisi, D.F. (2019) Pathogenetic, Prognostic, and Therapeutic Role of Fatty Acid Synthase in Human Hepatocellular Carcinoma. Frontiers in Oncology, 9, 1412. https://doi.org/10.3389/fonc.2019.01412
- 58. Amrutha, N.A., Retnakumari Archana, P., Mohan, S.G., John Anto, R. and Sadasivan, C. (2019) Pyridine Derivatives as Anticancer Lead Compounds with Fatty Acid Synthase as the Target: An in Silico-Guided in Vitro Study. Journal of Cellular Biochemistry, 120, 16643-16657. https://doi.org/10.1002/jcb.28923
- 59. Zhou, Y.Y., Su, W.P., Liu, H., Chen, T.L., Höti, N., Pei, H.P., et al. (2020) Fatty Acid Synthase Is a Prognostic Marker and Associated with Immune Infiltrating in Gastric Cancers Precision Medi-cine. Biomarkers in Medicine, 14, 185-199. https://doi.org/10.2217/bmm-2019-0476
- 60. Fhu, C.W. and Azhar, A. (2020) Fatty Acid Synthase: An Emerging Target in Cancer. Molecules, 25, 3935. https://doi.org/10.3390/molecules25173935
- 61. Rae, C., Haberkorn, U., Babich John, W. and Mairs, R.J. (2015) Inhibition of Fatty Acid Synthase Sensitizes Prostate Cancer Cells to Radiotherapy. Radiation Research, 184, 482-493. https://doi.org/10.1667/RR14173.1
- 62. Lei, T., Zhu, Y.T., Jiang, C.F., Wang, Y., Fu, J.F., Fan, Z., et al. (2016) MicroRNA-320 Was Downregulated in Non-Small Cell Lung Cancer and Inhibited Cell Proliferation, Migration and In-vasion by Targeting Fatty Acid Synthase. Molecular Medicine Reports, 14, 1255-1262. https://doi.org/10.3892/mmr.2016.5370
- 63. 颜新建, 李高峰, 唐梅, 杨小平. 下调脂肪酸合成酶表达对膀胱癌UMUC3细胞增殖、迁移及侵袭能力的影响[J]. 中国应用生理学杂志, 2019, 35(6): 543-547+476.
- 64. Li, X.X., Li, J.H., Jun, Q.M., Liu, J.K. and Hasim, A. (2020) SIRT3 Promotes the Invasion and Metastasis of Cervical Cancer Cells by Regulating Fatty Acid Synthase. Molecular and Cellular Biochemistry, 464, 11-20. https://doi.org/10.1007/s11010-019-03644-2
- 65. Quintela-Fandino, B. (2020) Emerging Role of Fatty Acid Syn-thase in Tumor Initiation: Implications for Cancer Prevention. Molecular & Cellular Oncology, 7, Article ID: 1709389. https://doi.org/10.1080/23723556.2019.1709389
- 66. De Piano, M., Manuelli, V., Zadra, G., Loda, M., Muir, G., Chandra, A., et al. (2020) Exploring a Role for Fatty Acid Synthase in Prostate Cancer Cell Migration. Small GTPases, 1-8. https://doi.org/10.1080/21541248.2020.1826781
- 67. Tongluan, N., Ramphan, S., Wintachai, P., Jaresit-thikunchai, J., Khongwichit, S., Wikan, N., et al. (2017) Involvement of Fatty Acid Synthase in Dengue Virus Infection. Virology Journal, 14, Article No. 28. https://doi.org/10.1186/s12985-017-0685-9
- 68. Zhang, N., Zhao, H.J. and Zhang, L.L. (2019) Fatty Acid Syn-thase Promotes the Palmitoylation of Chikungunya Virus nsP1. Journal of Virology, 93, e01747-18. https://doi.org/10.1128/JVI.01747-18
- 69. 陈哲, 周伊, 金宏威, 刘振明, 杨振军, 岳剑波, 张亮仁, 李汉璋, 张礼和. 钙信号相关多功能酶CD38的抑制剂研究[C]//中国药学会. 2011年全国药物化学学术会议——药物的源头创新论文摘要集. 中国药学会, 2011: 1.
- 70. Jia, B.L., Cheong, G.-W. and Zhang, S.H. (2013) Multifunctional Enzymes in Archaea: Promiscuity and Moonlight. Extremophiles, 17, 193-203. https://doi.org/10.1007/s00792-012-0509-1
- 71. Lorenzen, W., Ahrendt, T., Bozhüyük Kenan, A.J. and Bode, H.B. (2014) A Multifunctional Enzyme Is Involved in Bacterial Ether Lipid Biosynthesis. Nature Chemical Biology, 10, 425-427. https://doi.org/10.1038/nchembio.1526
- 72. Ibrahim, N.A., Abdel-Aziz, M.S., Eid, B.M. Hamdy, S.M. and Abdallah, S.E. (2016) Biosynthesis, Optimization and Potential Textile Application of Fungal Cellulases/Xylanase Multifunctional Enzyme Preparation from Penicillium sp. SAF6. Biocatalysis and Biotransformation, 34, 128-136. https://doi.org/10.1080/10242422.2016.1237943
- 73. Maier, T. (2017) Fatty Acid Synthases: Re-Engineering Biofactories. Nature Chemical Biology, 13, 344-345. https://doi.org/10.1038/nchembio.2338
- 74. Gao, L.Z., Fan, K.L. and Yan, X.Y. (2017) Iron Oxide Nanozyme: A Multifunctional Enzyme Mimetic for Biomedical Applications. Theranostics, 7, 3207-3227. https://doi.org/10.7150/thno.19738
- 75. Yarbrough John, M., Zhang, R.R., Mittal, A., Vander Wall, T., Bomble, Y.J., Decker, S.R., et al. (2017) Multifunctional Cellulolytic Enzymes Outperform Processive Fungal Cellulases for Coproduction of Nanocellulose and Biofuels. ACS Nano, 11, 3101-3109.
- 76. Chen, L., et al. (2020) Cellular Lipid Production by the Fatty Acid Synthase-Duplicated Lipomyces kononenkoae BF1S57 Strain for Biodiesel Making. Re-newable Energy, 151, 707-714. https://doi.org/10.1016/j.renene.2019.11.074
- 77. Suburu, J., Shi, L.H., Wu, J.S., Wang, S.H., Samuel, M., Thomas, M.J., et al. (2014) Fatty Acid Synthase Is Required for Mammary Gland Develop-ment and Milk Production during Lactation. American Journal of Physiology, Endocrinology and Metabolism, 306, E1132-E1143. https://doi.org/10.1152/ajpendo.00514.2013
- 78. 梁丽英, 张国海, 许瑞安. 脂肪酸合成酶抑制剂研究热点及其开发[J]. 中国现代应用药学, 2010, 27(4): 300-303.
- 79. Scott, M.J., Palmer, J.C., Tayler, H., Palmer, L.E., Ashby, E., Kehoe, P.G. and Love, S. (2014) Aβ Degradation or Cerebral Perfusion? Divergent Effects of Multifunctional Enzymes. Frontiers in Aging Neuroscience, 6, 238. https://doi.org/10.3389/fnagi.2014.00238
- 80. Narita, T.B, Kikukawa, T.W, Sato, Y.G., Miyazaki, S.H., Morita, N. and Saito, T. (2014) Role of Fatty Acid Synthase in the Development of Dictyostelium Discoideum. Journal of Oleo Science, 63, 281-289.
- 81. Singh, N., Manhas, A., Kaur, G., Jagavelu, K. and Hanif, K. (2016) Inhibition of Fatty Acid Synthase Is Protective in Pulmonary Hypertension. British Journal of Pharmacology, 173, 2030-2045. https://doi.org/10.1111/bph.13495
- 82. Neetu, S., Himalaya, S., Kumaravelu, J., Wahajuddin, M. and Hanif, K. (2017) Fatty Acid Synthase Modulates Proliferation, Metabolic Functions and Angiogenesis in Hypoxic Pulmonary Ar-tery Endothelial Cells. European Journal of Pharmacology, 815, 462-469. https://doi.org/10.1016/j.ejphar.2017.09.042
- 83. Fiedler, J.D, Fishman, M.R, Brown, S.D, Lau, J. and Finn, M.G. (2018) Multifunctional Enzyme Packaging and Catalysis in the Qβ Protein Nanoparticle. Biomacromolecules, 19, 3945-3957. https://doi.org/10.1021/acs.biomac.8b00885
- 84. Singh, N., Shafiq, M., Jagavelu, K. and Hanif, K. (2019) Involvement of Fatty Acid Synthase in Right Ventricle Dysfunction in Pulmonary Hypertension. Experimental Cell Research, 383, Arrticle ID: 111569. https://doi.org/10.1016/j.yexcr.2019.111569
- 85. 龚绍明, 何大乾, 沈洪民, 郁怀丹, 孙国荣, 许洪泉, 李庆荣, 刘军, 胡冲云. 多功能酶、福乐兴211、食益佳在肉鸭日粮中的应用试验[J]. 上海畜牧兽医通讯, 2005(5): 43-45.
- 86. 杨雪梅, 李智, 顾以韧, 陶璇, 梁艳, 杨跃奎, 钟志君, 陈晓晖, 吕学斌. 脂肪酸合成酶与不同猪种肌内脂肪含量关系研究[J]. 养猪, 2016(3): 60-61.
- 87. 唐晓伟, 刁青云. 蜜蜂体内多功能氧化酶诱导作为检测环境毒物的标准[J]. 中国蜂业, 2011, 62(Z2): 55-57.
- 88. Shelomi, M., Wipfler, B., Zhou, X. and Pauchet, Y. (2020) Multifunctional Cellulose Enzymes Are Ancestral in Polyneoptera. Insect Molecular Biology, 29, 124-135. https://doi.org/10.1111/imb.12614
- 89. Wang, S. and Yao, X. (2009) Diversity Analysis on Imbalanced Data Sets by Using Ensemble Models. 2009 IEEE Symposium on Computational Intelligence and Data Mining, Nashville, 30 March-2 April 2009, 324-331. https://doi.org/10.1109/CIDM.2009.4938667
