Pharmacy Information
Vol.
12
No.
05
(
2023
), Article ID:
72139
,
9
pages
10.12677/PI.2023.125051
靶向p53-MDM2的喹啉类抗肿瘤小分子化合物的虚拟筛选
丁涵静1,胡芳2,胡美纯1*
1湖北科技学院基础医学院,湖北 咸宁
2咸宁波力体育文化有限公司,湖北 咸宁
收稿日期:2023年8月4日;录用日期:2023年9月1日;发布日期:2023年9月11日

摘要
p53能诱导细胞凋亡,抑制肿瘤的发生发展。MDM2是p53最重要的负调控因子,能诱导p53的泛素化降解,抑制p53的功能。因此,针对p53-MDM2设计小分子靶向抑制剂是开发新型抗肿瘤药物的一种极具前景的策略。喹啉类化合物及其衍生物是具有抗肿瘤活性的小分子药物的重要来源。本研究以p53-MDM2为药物靶点,对喹啉类化合物库进行了基于药效团和分子对接的虚拟筛选,最终获得了具有潜在的靶向p53-MDM2抗肿瘤的小分子化合物Compound 6和Compound 9。本研究为靶向抗癌药物研发提供了一种筛选方法,并为进一步开发针对p53-MDM2的小分子靶向药奠定了基础。
关键词
p53,MDM2,抗肿瘤小分子化合物,喹啉,虚拟筛选

Virtual Screening of Quinoline Antitumor Small Molecular Compounds Targeting the p53-MDM2 Interaction
Hanjing Ding1, Fang Hu2, Meichun Hu1*
1School of Basic Medical Sciences, Hubei University of Science and Technology, Xianning Hubei
2Xianning Bo-li Sports Culture Co., Ltd., Xianning Hubei
Received: Aug. 4th, 2023; accepted: Sep. 1st, 2023; published: Sep. 11th, 2023

ABSTRACT
p53 can induce cell apoptosis and inhibit tumor occurrence and development. MDM2 is the most important negative regulator of p53, which can induce the ubiquity nation and degradation of p53, and inhibit its function. Therefore, designing small molecular inhibitors against p53-MDM2 is a promising strategy for the development of novel antitumor drugs. Quino line and its derivatives are important sources of small molecular drugs with potential antitumor activities. In this work, using the p53-MDM2 as the target, we performed virtual screening on the quino line compound library based on pharmacophore modeling and molecular docking, and finally obtained Compound 6 and Compound 9 which have potential antitumor activities against p53-MDM2.This work provides a screening method for the discovery of targeted antitumor drugs, and lays the foundation for further development of small molecular inhibitors against p53-MDM2.
Keywords:p53, MDM2, Antitumor Small Molecular Compounds, Quinoline, Virtual Screening

Copyright © 2023 by author(s) and Hans Publishers Inc.
This work is licensed under the Creative Commons Attribution International License (CC BY 4.0).
http://creativecommons.org/licenses/by/4.0/


1. 引言
p53蛋白是由抑癌基因TP53编码的转录因子,可通过调控多个下游靶基因的表达,实现细胞凋亡、细胞周期停滞、DNA修复等功能,从而抑制肿瘤发生发展 [1] 。MDM2蛋白(Murine Double Minute 2)是p53的负调控因子,具有E3泛素连接酶活性,能促进p53的泛素化,使p53被蛋白酶体识别并降解 [2] 。如果阻断p53与MDM2之间的相互作用,就能提高p53蛋白的稳定性,使p53得以持续发挥其抑癌功能,这也是目前肿瘤靶向治疗的重要策略之一 [2] [3] 。在这一策略的基础上,研究人员已开发出不少靶向p53-MDM2的小分子药物,其中部分药物已进入临床试验阶段。例如,RG7112就是一种能有效抑制p53与MDM2结合的小分子化合物 [3] ,是首批进入临床试验的p53-MDM2抑制剂之一,且已完成了针对各种不同癌症类型的Ⅰ期临床研究(NCT00623870, NCT00559533, NCT01164033)。目前已进入临床试验的p53-MDM2小分子抑制剂还有AMG232、RG7388、APG115、HDM201等等。正因为p53-MDM2这一靶点对于肿瘤治疗具有重要意义,所以其小分子靶向抑制剂的临床研究也在不断增加。然而,虽然现在已发现不少能抑制p53-MDM2的小分子药物,但能通过临床试验筛选的药物极少 [4] [5] ,目前暂无相关药物上市,迫切需要开发新型小分子靶向药。
喹啉类化合物泛指结构中含喹啉或异喹啉基团的小分子化合物,在自然界中广泛存在,在抗肿瘤药物开发方面具有很大的潜力 [6] [7] [8] [9] 。不少天然或者合成的喹啉类小分子均已有报道具有显著的抗癌作用,如甲基莲心碱、石蒜碱、蝙蝠葛碱、血根碱和小檗碱等等 [10] - [15] 。例如,本研究团队曾发现,石蒜碱能靶向EGFR抑制前列腺癌及神经胶质瘤细胞的增殖,诱导肿瘤细胞凋亡,且其抑癌机制与EGFR诱导的JAK/STAT信号通路有关 [10] [16] 。另外,已有多项研究报道,喹啉类化合物的抗癌机制可能与诱导细胞周期停滞、细胞凋亡、抑制血管生成和破坏细胞迁移有关 [17] - [21] 。因此,开发喹啉类小分子化合物作为抗肿瘤的候选药物具有极佳的应用前景。
虽然目前已出现许多有潜力的靶向p53-MDM2喹啉类小分子药物,且其体外或体内抗肿瘤活性也已得到了验证;但是,体外实验的筛选效率较低,研发周期较长、成本较大。若采用计算机辅助药物设计的方法,就能够降低实验成本、缩短研发周期、提高药物研发效率,目前这已成为高通量筛选并获得高活性候选药物的重要手段 [22] [23] 。本研究拟利用基于简单药效团及分子对接的虚拟筛选方法,对靶向p53-MDM2的喹啉醇类小分子抑制剂进行研究分析,探索小分子化合物与MDM2的结合方式,最终筛选出能抑制p53-MDM2的新型候选先导化合物,为进一步研究喹啉类小分子化合物的抗肿瘤活性奠定基础。
2. 方法
2.1. 小分子配体的准备
将本实验室自有的82个喹啉类化合物及其衍生物的分子结构以mol2格式保存,运行SYBYL2.0软件并导入自主构建的小分子化合物,再对化合物进行几何优化与力场优化等操作,将所有小分子结构以PDBQT格式保存,完成小分子化合物库的建立。
2.2. 蛋白质靶点的准备
在PDB数据库(ProteinDataBank)中搜集分辨率较高的MDM2蛋白质晶体结构,导入SYBYL2.0软件,删除水分子和原配体,添加缺失残基,再进行加氢、加电荷处理,计算残基质子化状态和氢键网络,确定对接的活性位点,再将所有蛋白质结构转换为PDBQT格式并保存,随后与晶体结构中的小分子配体进行初步的交叉对接以确定适用于虚拟筛选的最佳MDM2蛋白质结构。
2.3. 基于药效团模型和分子对接的虚拟筛选
本研究整合了两种常用的药物筛选策略(图1),先进行基于药效团模型的批量筛选,再进行基于分子对接的精细筛选,以期筛选出最优候选先导化合物。首先,基于MDM2与p53肽段的蛋白质共结晶结构及MDM2与7种非肽小分子抑制剂的共结晶结构,开发一个简单的药效团模型。该模型用于执行药效团筛选,整理出符合该模型的化合物。由于药效团搜索显著减少了用于下一步分子对接研究的化合物数量,因此可以采用更耗时、更精细的分子对接来获得更准确的配体结合模型。然后,采用Autodock软件对候选化合物与MDM2进行精细对接,获得小分子与MDM2的10个结合构象,最后根据亲和力及RMSD值评估最佳结合构象并进行可视化分析。

Figure 1. An integrated computational screening strategy for the small molecular lead compounds targeting the p53-MDM2 interaction
图1. 靶向p53-MDM2的小分子先导化合物的计算机综合筛选策略
3. 结果与讨论
3.1. MDM2蛋白质结构的选择
在PDB数据库中搜集了7个分辨率较高(≤2.3 Å)的MDM2与小分子配体的共结晶复合物结构,将原小分子配体与蛋白质分离,并进行初步的交叉对接,并与原复合物结晶结构进行比对(表1)。结果显示,在本研究中采用的计算策略背景下,PDB编号为3JZK的蛋白质结构的对接性能表现最佳,平均RMSD值最低,对小分子配体结合构象的预测最准。如图2所示,经三次平行对接后,可见Dockingmode 1和Dockingmode 2的小分子配体预测构象与原配体近乎一致。因此,在本研究的后续工作中均采用编号为3JZK的MDM2晶体结构。
Table 1. Cocryst allized structures of MDM2 and its ligands and the average cross-docking RMSD values
表1. MDM2与其小分子配体的共结晶结构及交叉对接的平均RMSD值
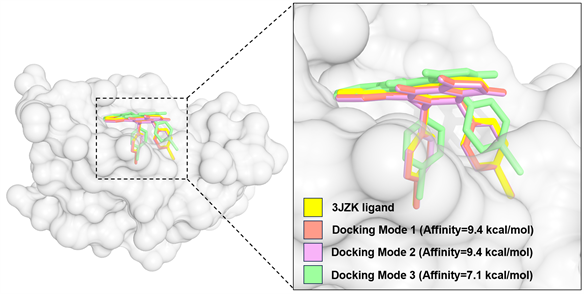
Figure 2. Superimposed binding modes of a ligand docked to MDM2 (PDB code: 3JZK).
图2. 小分子配体对接MDM2的结合模式叠合图(PDB code: 3JZK)
3.2. 蛋白质靶点结构分析及药效团模型构建
基于药效团的筛选和基于结构的筛选是两种常用的计算机辅助虚拟筛选技术。基于药效团的筛选方法能够快速淘汰化合物,但其缺点是对空间匹配度的考虑不足。基于结构的筛选方法能够计算小分子配体与蛋白质结合的亲和力,但缺点是在计算资源有限的情况下可能会导致对结合构象的预测不准 [24] [25] [26] 。为了充分利用两种方法的优势,互相弥补不足,本研究采用了一种综合的策略(图1):首先,对MDM2与已知的小分子抑制剂的结合模式进行分析,再根据分析结果构建药效团模型,该模型用于筛选出符合条件的小分子化合物;然后,再用基于结构的筛选将小分子化合物对接到MDM2的p53结合口袋,并根据亲和力的大小对其进行排序。由于药效团筛选能大幅度降低小分子化合物的数量,后续就可以采用更精细的对接程序,得到更准确的蛋白质和配体的结合模型。
对MDM2与其小分子抑制剂共结晶的结构分析表明,所有7种已知的MDM2小分子抑制剂均能较好地模拟p53与MDM2的“结合热点”残基,即p53的三个疏水残基Phe19、Trp23、Leu26 (图3A)。对已知小分子配体进行结构叠合分析结果表明,所有已知小分子抑制剂均能与以上p53的三个残基结合亚口袋相互作用,且三个疏水核心之间的距离分别为:5.6 ± 1.0Å (Phe19-Trp23)、5.1 ± 1.2Å (Phe19-Leu26)、7.2 ± 1.5Å (Trp23-Leu26) (图3B)。根据7个小分子配体的结构及其与MDM2的相互作用模式,开发一个简单的药效团模型(图4)。该模型的组成是H1H2H3H4 (H为Hydrophobic),其中H1和H2的空间分布较为分散,而H3与H4的距离较为接近(<4Å),H1、H2、H3-H4分别对应于MDM2的三个疏水结合亚口袋。
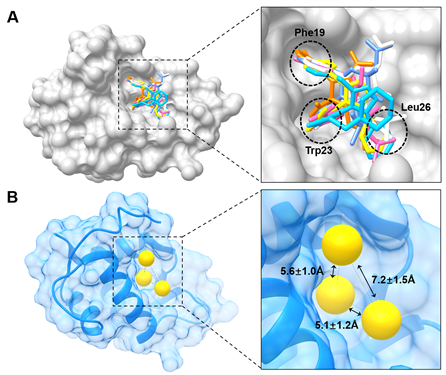
Figure 3. Analyses of thecocry stallized structures of MDM2 and its small ligands. (A) Superimposition of MDM2 ligands; (B) Relative locations of the three hydrophobic binding subpockets of MDM2
图3. MDM2与其小分子配体共结晶的结构分析。(A) MDM2与小分子配体的共结晶结构叠合图;(B) MDM2的三个疏水残基结合亚口袋的相对位置图

Figure 4. Pharmacophore hypothesis and characteristics for the virtual screening of this work
图4. 用于虚拟筛选的药效团模型及特征
3.3. 基于药效团及分子对接的虚拟筛选
第一轮筛选采用基于药效团的虚拟筛选,从化合物库中得到了9个符合规则的喹啉醇类小分子化合物(图5)。结构叠合分析结果显示,9个小分子化合物与药效团模型HHHH高度匹配(图6A)。由于基于配体的药效团模型缺乏蛋白质柔性结构的分析信息,因此下一轮筛选的方法采用基于分子对接的虚拟筛选。第二轮筛选采用Autodock将小分子化合物对接到MDM2的疏水口袋中,以已被广泛使用的靶向p53-MDM2小分子药物Nutlin3a [27] 为阳性对照。经对接后,每个小分子得到10个结合构象,再根据Affinity值及RMSD值进行综合评价打分,分析和比较每个小分子与MDM2的匹配度(表2)。结果显示,Compound 6和Compound 9的Affinity分别为−9.1kcal/mol及−9.8kcal/mol,而阳性对照Nutlin3a为−8.3kcal/mol;因此,Compound 6和Compound 9的亲和力均优于Nutlin3a。结合模式分析结果显示,Compound 6和Compound 9均能与MDM2的三个结合亚口袋相互作用(表2及图6B)。
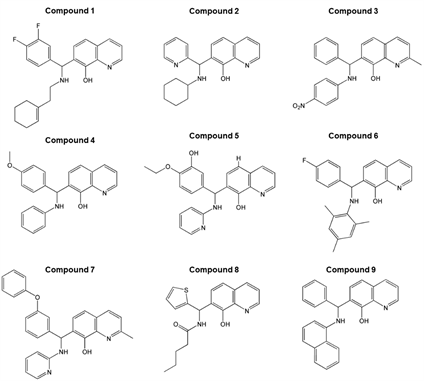
Figure 5. Structures of the nine small compound candidates
图5. 九个候选小分子化合物分子结构
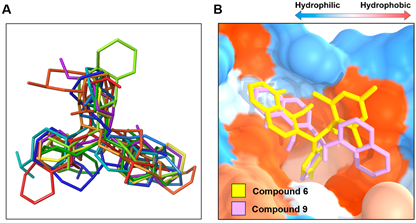
Figure 6. Superimposition of the structures of small compound candidates.(A) Superimposition of the nine small molecule compound candidates after the first screening; (B) Superimposition of the two small molecular compounds docked to the MDM2 hydrophobic pocket after the second screening
图6. 候选小分子化合物结构叠合图。(A) 第一轮筛选得到的九个候选小分子化合物叠合图;(B) 第二轮筛选得到的两个小分子化合物对接MDM2疏水口袋的叠合图
Table 2. Affinity and RMSD values from the optimal binding mode of the small molecule candidate compound sbymolecular docking
表2. 候选小分子化合物对接MDM2最优结合构象的亲和力及RMSD值
a. Root mean square deviation lower bound;
b. Root mean square deviation upper bound。
PubChem数据库搜索结果显示,Compound 6和Compound 9均未有以p53-MDM2为靶点的抗肿瘤研究报道,说明本研究筛选出的化合物具有结构新颖性。文献搜索结果显示,与本研究的候选化合物较为相似的一种喹啉醇类化合物在体外实验中能抑制Rce1蛋白酶的功能,且细胞毒性较低,具有较好的靶向Rce1抗结肠癌的活性 [28] 。因此,本研究得到的候选化合物极有可能也具有潜在的抗肿瘤活性。Compound 6和Compound 9的后续抗肿瘤体外研究正在进行中。
4. 结论
近年来,随着肿瘤发病率的不断攀升,迫切需要开发新型抗肿瘤药物。p53-MDM2是肿瘤治疗的关键靶点,抑制p53-MDM2相互作用能够延续p53的抑癌功能,从而抑制肿瘤生长。针对该靶点,已有各类p53-MDM2的抑制剂被开发出来,包括抗体类、肽类、小分子类等等。其中,小分子类抑制剂具备可口服、渗透率高,且储存运输成本较低的特性,已成为研发的热点。本研究根据p53与MDM2的相互作用结构特征及MDM2与7个已知小分子抑制剂的相互作用模式构建了一个药效团模型。运用该模型对化合物库进行第一轮筛选结果显示,有9个喹啉醇类小分子的匹配度高,符合药效团特征。采用分子对接进行第二轮筛选的结果显示,Compound 6和Compound 9的亲和力较高,且优于已知的靶向MDM2的抗肿瘤药物Nutlin3a。结合模式分析显示,Compound 6和Compound 9均能与MDM2的疏水口袋相互作用,且主要相互作用力为疏水堆积力。PubChem数据库的搜索结果显示,本研究筛选出的候选化合物均未见报道,而相似度较高的另一个喹啉醇化合物表现出了显著的抗肿瘤活性。因此,本研究得出的候选化合物极有可能具备靶向p53-MDM2抑制肿瘤细胞生长的潜力。综上所述,本研究为开发靶向p53-MDM2的新型抗肿瘤抑制剂提供了筛选方法,并获得了两个有潜力的候选化合物,为将来进一步开发和验证喹啉醇类的小分子抗肿瘤化合物奠定了理论基础。
基金项目
咸宁市自然科学基金项目(2022ZRKX066),湖北科技学院国家培育项目(2022-24GP02),湖北省自然科学基金面上项目(2022CFB394),湖北省卫生健康委员会科研项目(WJ2019Q022),湖北科技学院医学科研专项基金(2022YKY08),湖北科技学院横向科研项目(2022HX206)。
文章引用
丁涵静,胡芳,胡美纯. 靶向p53-MDM2的喹啉类抗肿瘤小分子化合物的虚拟筛选
Virtual Screening of Quinoline Antitumor Small Molecular Compounds Targeting the p53-MDM2 Interaction[J]. 药物资讯, 2023, 12(05): 434-442. https://doi.org/10.12677/PI.2023.125051
参考文献
- 1. Kastenhuber, E.R. and Lowe, S.W. (2017) Putting p53 in Context, Cell, 170, 1062-1078. https://doi.org/10.1016/j.cell.2017.08.028
- 2. Bhatia, N., Khator, R., Kulkarni, S., Singh, Y., Kumar, P. and Tha-reja, S. (2023) Recent Advancements in the Discovery of MDM2/MDM2-p53 Interaction Inhibitors for the Treatment of Cancer. Current Medicinal Chemistry, 30, 3668-3701. https://doi.org/10.2174/0929867330666221114103924
- 3. Chahat, Bhatia, R. and Kumar, B. (2023) P53 as a Po-tential Target for treatment of Cancer: A Perspective on Recent Advancements in Small Molecules with Structural In-sights and SAR Studies. European Journal of Medicinal Chemistry, 247, Article ID: 115020. https://doi.org/10.1016/j.ejmech.2022.115020
- 4. Zhu, H., Gao, H., Ji, Y., Zhou, Q., Du, Z., Tian, L., Jiang, Y., Yao, K. and Zhou, Z. (2022) Targeting p53-MDM2 Interaction by Small-Molecule Inhibitors: Learning from MDM2 In-hibitors in Clinical Trials. Journal of Hematology & Oncology, 15, Article No. 91. https://doi.org/10.1186/s13045-022-01314-3
- 5. Nishikawa, S. and Iwakuma, T. (2023) Drugs Targeting p53 Mutations with FDA Approval and in Clinical Trials. Cancers, 15, Article No. 429. https://doi.org/10.3390/cancers15020429
- 6. Afzal, O., Kumar, S., Haider, M.R., Ali, M.R., Kumar, R., Jaggi, M. and Bawa, S. (2015) A Review on Anticancer Potential of Bioactive Heterocycle Quinoline. European Journal of Me-dicinal Chemistry, 97, 871-910. https://doi.org/10.1016/j.ejmech.2014.07.044
- 7. Lauria, A., La Monica, G., Bono, A. and Martorana, A. (2021) Quinoline Anticancer Agents Active on DNA and DNA-Interacting Proteins: From Classical to Emerging Therapeutic Targets. European Journal of Medicinal Chemistry, 220, Article ID: 113555. https://doi.org/10.1016/j.ejmech.2021.113555
- 8. Liu, K., Mo, M., Yu, G., Yu, J., Song, S.M., Cheng, S., Li, H.M., Meng, X.L., Zeng, X.P., Xu, G.C., et al. (2023) Discovery of Novel 2-(Trifluoromethyl)Quinolin-4-Amine De-rivatives as Potent Antitumor Agents with Microtubule Polymerization Inhibitory Activity. Bioorganic Chemistry, 139, Article ID: 106727. https://doi.org/10.1016/j.bioorg.2023.106727
- 9. Ilovaisky, A.I., Scherbakov, A.M., Merkulova, V.M., Cherno-burova, E.I., Shchetinina, M.A., Andreeva, O.E., Salnikova, D.I., Zavarzin, I.V. and Terent'ev, A.O. (2023) Secoster-oid-Quinoline Hybrids as New Anticancer Agents. The Journal of Steroid Biochemistry and Molecular Biology, 228, Article ID: 106245. https://doi.org/10.1016/j.jsbmb.2022.106245
- 10. Shen, J., Zhang, T., Cheng, Z., Zhu, N., Wang, H., Lin, L., Wang, Z., Yi, H. and Hu, M. (2018) Lycorine Inhibits Glioblastoma Multiforme Growth through EGFR Suppression. Journal of Experimental & Clinical Cancer Research, 37, Article No. 157. https://doi.org/10.1186/s13046-018-0785-4
- 11. Zhang, Y.-B., Fei, H.-X., Guo, J., Zhang, X.-J., Wu, S.-L. and Zhong, L.-L. (2019) Dauricine Suppresses the Growth of Pancreatic Cancer in Vivo by Modulating the Hedgehog Sig-naling Pathway. Oncology Letters, 18, 4403-4414. https://doi.org/10.3892/ol.2019.10790
- 12. Li, S., Zhang, Y., Zhang, J., Yu, B., Wang, W., Jia, B., Chang, J. and Liu, J. (2022) Neferine Exerts Ferroptosis-Inducing Effect and Antitumor Effect on Thyroid Cancer through Nrf2/HO-1/NQO1 Inhibition. Journal of Oncology, 2022, Article ID: 7933775. https://doi.org/10.1155/2022/7933775
- 13. Sun, Y., Gu, Y., Gao, X., Jin, X., Wink, M., Sharopov, F.S., Yang, L. and Sethi, G. (2023) Lycorine Suppresses the Malignancy of Breast Carcinoma by Modulating Epithelial Mesenchymal Transition and β-Catenin Signaling. Pharmacological Research, 195, Article ID: 106866. https://doi.org/10.1016/j.phrs.2023.106866
- 14. Wang, K.D., Zhu, M.L., Qin, C.J., Dong, R.F., Xiao, C.M., Lin, Q., Wei, R.Y., He, X.Y., Zang, X., Kong, L.Y., et al. (2023) Sanguinarine Induces Apoptosis in Osteosarcoma by At-tenuating the Binding of STAT3 to the Single-Stranded DNA-Binding Protein 1 (SSBP1) Promoter Region. British Journal of Pharmacology. https://doi.org/10.1111/bph.16202
- 15. Chen, Z., Dong, Y., Yan, Q., Li, Q., Yu, C., Lai, Y., Tan, J., Fan, M., Xu, C., Li, L., et al. (2023) Liquid Chromatography-Tandem Mass Spectrometry Analysis of a Ratio-Optimized Drug Pair of Sophora flavescens Aiton and Coptis chinensis Franch and Study on the Mechanism of Anti-Colorectal Cancer Effect of Two Alkaloids Thereof. Frontiers in Oncology, 13, Article 1198467. https://doi.org/10.3389/fonc.2023.1198467
- 16. Hu, M., Peng, S., He, Y., Qin, M., Cong, X., Xing, Y., Liu, M. and Yi, Z. (2015) Lycorine Is a Novel Inhibitor of the Growth and Metastasis of Hormone-Refractory Prostate Cancer. Oncotarget, 6, 15348-15361. https://doi.org/10.18632/oncotarget.3610
- 17. Solomon, V.R. and Lee, H. (2011) Quinoline as a Privileged Scaf-fold in Cancer Drug Discovery. Current Medicinal Chemistry, 18, 1488-1508. https://doi.org/10.2174/092986711795328382
- 18. Man, R.J., Jeelani, N., Zhou, C. and Yang, Y.S. (2021) Recent Progress in the Development of Quinoline Derivatives for the Exploitation of Anti-Cancer Agents. Anti-Cancer Agents in Medicinal Chemistry, 21, 825-838. https://doi.org/10.2174/1871520620666200516150345
- 19. Pradhan, V., Salahuddin, Kumar, R., Mazumder, A., Abdullah, M.M., Shahar Yar, M., Ahsan, M.J. and Ullah, Z. (2023) Molecular Target Interactions of Quinoline Deriva-tives as Anticancer Agents: A Review. Chemical Biology & Drug Design, 101, 977-997. https://doi.org/10.1111/cbdd.14196
- 20. Sonawane, H.R., Vibhute, B.T., Aghav, B.D., Deore, J.V. and Patil, S.K. (2023) Versatile Applications of Transition Metal Incorporating Quinoline Schiff Base Metal Complexes: An Overview. European Journal of Medicinal Chemistry, 258, Article ID: 115549. https://doi.org/10.1016/j.ejmech.2023.115549
- 21. Tyagi, S., Salahuddin, Mazumder, A., Kumar, R., Datt, V., Shabana, K., Yar, M.S. and Ahsan, M.J. (2023) Synthesis and SAR of Potential Anti-Cancer Agents of Quinoline Ana-logues: A Review. Medicinal Chemistry, 9, 785-812. https://doi.org/10.2174/1573406419666230228140619
- 22. de Araújo, R.S.A., da Silva-Junior, E.F., de Aquino, T.M., Scotti, M.T., Ishiki, H.M., Scotti, L. and Mendonça-Junior, F.J.B. (2020) Computer-Aided Drug Design Applied to Secondary Metabolites as Anticancer Agents. Current Topics in Medicinal Chemistry, 20, 1677-1703. https://doi.org/10.2174/1568026620666200607191838
- 23. Vemula, D., Jayasurya, P., Sushmitha, V., Kumar, Y.N. and Bhandari, V. (2023) CADD, AI and ML in Drug Discovery: A Comprehensive Review. European Journal of Pharmaceutical Sciences, 181, Article ID: 106324. https://doi.org/10.1016/j.ejps.2022.106324
- 24. Abdolmaleki, A., Ghasemi, J.B. and Ghasemi, F. (2017) Computer Aided Drug Design for Multi-Target Drug Design: SAR /QSAR, Molecular Docking and Pharmacophore Methods. Current Drug Targets, 18, 556-575. https://doi.org/10.2174/1389450117666160101120822
- 25. Dar, K.B., Bhat, A.H., Amin, S., Hamid, R., Anees, S., Anjum, S., Reshi, B.A., Zargar, M.A., Masood, A. and Ganie, S.A. (2018) Modern Computational Strategies for De-signing Drugs to Curb Human Diseases: A Prospect. Current Topics in Medicinal Chemistry, 18, 2702-2719. https://doi.org/10.2174/1568026619666190119150741
- 26. Giordano, D., Biancaniello, C., Argenio, M.A. and Facchiano, A. (2022) Drug Design by Pharmacophore and Virtual Screening Approach. Pharmaceuticals, 15, Article No. 646. https://doi.org/10.3390/ph15050646
- 27. Haronikova, L., Bonczek, O., Zatloukalova, P., Kokas-Zavadil, F., Kucerikova, M., Coates, P.J., Fahraeus, R. and Vojtesek, B. (2021) Resistance Mechanisms to Inhibitors of p53-MDM2 Interactions in Cancer Therapy: Can We Overcome Them? Cellular & Molecular Biology Letters, 26, Article No. 53. https://doi.org/10.1186/s11658-021-00293-6
- 28. Mohammed, I., Hampton, S.E., Ashall, L., Hildebrandt, E.R., Kutlik, R.A., Manandhar, S.P., Floyd, B.J., Smith, H.E., Dozier, J.K., Distefano, M.D., et al. (2016) 8-Hydroxyquinoline-Based Inhibitors of the Rce1 Protease Disrupt Ras Membrane Localization in Human Cells. Bioor-ganic & Medicinal Chemistry, 24, 160-178. https://doi.org/10.1016/j.bmc.2015.11.043
NOTES
*通讯作者。
