Advances in Microbiology
Vol.
08
No.
03
(
2019
), Article ID:
32252
,
9
pages
10.12677/AMB.2019.83018
Advances in Bacterial Lantibiotics
Huafei Zhou, Hongfu Yang, Zhaolin Shu, Hongzhou Chen, Kebing Yao*
Huafei Zhou, Hongfu Yang, Zhaolin Shu, Hongzhou Chen, Kebing Yao*
Zhenjiang Academy of Agricultural Sciences in Hilly Areas of Jiangsu Province, Jurong Jiangsu

Received: Aug. 29th, 2019; accepted: Sep. 13th, 2019; published: Sep. 20th, 2019

ABSTRACT
Lantibiotics is a class of antimicrobial peptides, or thioether antibiotics produced by bacteria that are synthesized by the ribosomal pathway and has a special organic group. Lantibiotics is produced by Gram-positive bacteria, and can significantly inhibit the growth of microorganisms, especially Gram-positive bacteria. Excessive use of traditional antibiotics leads to the widespread occurrence of pathogen resistance, lantibiotics has great potential for changing this situation. This study is based on the mechanism of action and product development of lantibiotics, to introduce the introduction, classification, mechanism of action, industrialization process and future prospects of lantibiotics one by one.
Keywords:Lantibiotics, Gram-Positive Bacteria, Antimicrobial Peptide, Discovery, Classification, Mechanism of Action, Prospect
细菌羊毛硫抗生素研究进展
周华飞,杨红福,束兆林,陈宏州,姚克兵*
江苏丘陵地区镇江农业科学研究所,江苏 句容

收稿日期:2019年8月29日;录用日期:2019年9月13日;发布日期:2019年9月20日

摘 要
羊毛硫抗生素(lantibiotics)是指由细菌产生的,通过核糖体途径合成的翻译后修饰的具有特殊有机基团的一类抗菌肽,或称硫醚抗生素。羊毛硫抗生素由革兰氏阳性菌产生,可以明显抑制微生物的生长,尤其是抑制革兰氏阳性菌。传统抗生素的过量使用导致病原菌抗药性的现象普遍发生,羊毛硫抗生素对于改变这一现状具有很大的开发潜力。本文根据羊毛硫抗生素的作用机制与产品开发研究现状,对羊毛硫抗生素的发现、分类、作用机制、产业化进程及未来的展望逐一介绍。
关键词 :羊毛硫抗生素,革兰氏阳性菌,抗菌肽,发现,分类,作用机制,展望

Copyright © 2019 by author(s) and Hans Publishers Inc.
This work is licensed under the Creative Commons Attribution International License (CC BY).
http://creativecommons.org/licenses/by/4.0/


1. 引言
抗菌肽是生物体内经诱导产生的一类具有生物活性的小分子多肽,分子量在3~5 kDa之间,具有热稳定性及广谱抗菌特性 [1] [2] ,羊毛硫抗生素是一类作用于细胞膜上的新型抗菌肽,主要结构特点为含有非编码的翻译后修饰氨基酸–羊毛硫氨酸(Ala)、甲基羊毛硫氨酸、β-甲基羊毛硫氨酸(Aba)和脱氢丙氨酸等(Dha) [3] [4] [5] [6] ,大部分羊毛硫抗生素还包含其他的修饰残基或环状结构,例如美杀菌素(mersacidin)和表皮菌素(epidermin)中的2-硫乙胺基团 [7] [8] [9] ,肉桂霉素(cinnamycin)中的赖丙氨酸和羟基天冬氨酸 [10] [11] ,乳杆菌素(lacticin) S和乳杆菌素3147中的D-丙氨酸 [12] [13] ,Pep5和乳杆菌素在N端的氧代乙酰 [14] 及放线菌合成microbisporicin抗生素中的羟脯氨酸等 [15] [16] [17] (表1),此类抗生素近年来才逐渐被人们发现、了解并开发。目前的羊毛硫抗生素全部是由革兰氏阳性菌产生,对阳性菌产生抑制作用,对阴性菌无效,这主要是由于革兰氏阴性菌细胞外膜能有效阻止羊毛硫抗生素的渗透导致的 [18] 。
2. 羊毛硫抗生素的发现历史
1928年Rogers报道乳酸菌的代谢产物可以抑制乳酸杆菌及其他链球菌的生长,该代谢产物后来被证实为乳酸链球菌素(Nisin) [19] [20] ,首次报道了结构内部含有羊毛硫氨酸的这类物质,随后在1947年被Mattick和Hirsch从乳酸杆菌中成功分离出来。1951年Hirsch首先将乳酸链球菌素用作食品防腐剂抑制肉毒棱状芽胞杆菌引起的食品腐败问题,直到现在依旧是全球商品化最成功的羊毛硫抗生素。1988年Buchaman首次提出乳酸链球菌素的合成机制,指出其前体物质是由57个氨基酸组成,包括含有23个氨基酸的N端前导序列和34个氨基酸的C端,经过翻译后修饰才可以形成具有生物活性的大分子,并被Kaletta和Entian加以证实 [21] [22] 。同年又在葡萄球菌上发现的羊毛硫抗生素表皮菌素(Epidermin)被证实了其由结构基因epiA编码形成抗生素的表皮纤维,形成的产物是一个由52个氨基酸组成的前导肽,经过一系列的后修饰加工环节形成成熟的Epidermin分子 [23] ,再次推动对羊毛硫抗生素研究的大发展。
3. 羊毛硫抗生素的结构特点与分类
3.1. 羊毛硫抗生素的结构特点
羊毛硫抗生素最显著的区别于其他抗生素的特点为含有羊毛硫氨酸和β-甲基羊毛硫氨酸等具备双羧基和双氨基的稀有氨基酸 [24] ,还含有一些其他类型的后修饰氨基酸,如脱氢丙氨酸(Dha)和脱氢丁氨酸(Dhb)等,这些稀有氨基酸在羊毛硫抗生素结构内部以共价键的方式形成内环,使羊毛硫抗生素具备较好的稳定性。例如乳酸链球菌素Nisin (图1),由34个氨基酸组成,分子式是C143H228O37N42S7,其分子结构中包含5种稀有氨基酸即Aba、Dha、Dhb、Ala-S-Ala和Ala-S-Aba,它们通过硫醚键形成五个内环,其活性分子常为二聚体或四聚体,到现在为止人们共发现其A、B、C、D、E、Z构型(图2),其中以A与Z两种构型最为活跃,两者的差别仅为在氨基酸顺序上第27位的氨基酸不同,A构型为组氨酸(His),Z构型为天门冬酰胺(Asn),同等浓度下乳酸链球菌素Z在溶解度和杀菌能力上均比乳酸链球菌素A强 [25] [26] 。成熟的乳酸链球菌素分子顶端还具有一定的弯曲 [27] ,内部三维结构显示还存在一个柔性铰链区 [28] 。

Figure 1. Molecular structures of lantibiotics [29]
图1. 羊毛硫抗生素的分子结构 [29]
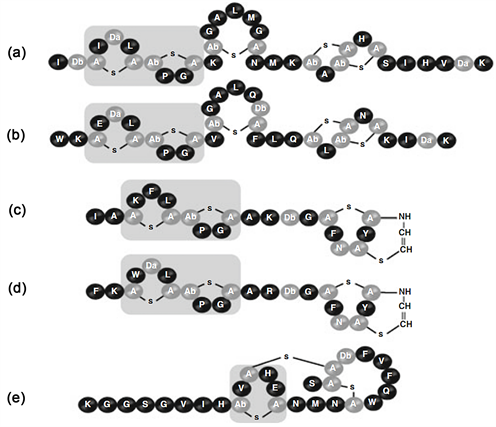
Figure 2. 5 configurations of Nisin [29]
图2. Nisin的5种构型 [29]
3.2. 羊毛硫抗生素的分类
依据羊毛硫抗生素分子结构与抑菌活性的不同,Jung [30] 建议将其分类A、B两类(如表1所示):A类为拉伸的线性螺旋状结构(如图1乳酸链球菌素所示),带有正电荷,具两亲性,能在细胞膜上形成孔洞致使细胞死亡,Asaduzzaman [31] 根据结构进一步把A类细分为拉伸的AⅠ型和具有N端线性区域和C端环形区域的AⅡ型;B类为球形带负电荷或者不带电荷,能抑制细胞壁的形成(如图1耐久霉素所示)。羊毛硫抗生素分类中还包括一个特殊的成员-AB双组份羊毛硫抗生素乳酸菌素(Lacticin) 3147,该抗生素由两个不同的多肽组成,同时具备A和B两类的特征,其中lanA1编码形成的肽为B类羊毛硫抗生素,lanA2编码的肽为A型羊毛硫抗生素,两者共同作用形成具备活性的羊毛硫抗生素 [32] 。A类中代表性的是乳酸链球菌素(Nisin)、枯草菌素(Subtilin)、表皮菌素(Epidermin)和Pep5,其中乳酸链球菌素的分子结构早在20世纪60~70年代就已经由Gross和Movell解析出来 [33] [34] ,其中脱氢丙氨酸和脱氢丁氨酸等非编码氨基酸的存在证实了乳酸链球菌素具有翻译后修饰过程。B类羊毛硫抗生素典型代表为美杀菌素(Mersacidine)、阿肽加定(Actagardin)和肉桂霉素(Cinnamycin) (图1)。美杀菌素是迄今为止分离出的最小的羊毛硫氨酸抗生素(1825Da,表1),是由20个氨基酸组成的四环肽,主要产生于芽孢杆菌对数生长后期,含有三个甲基羊毛硫氨酸残基、一个脱氢丙氨酸和一个S一氨基乙烯基-2-甲基半胱氨酸,不带净电荷,具疏水性。在皮下注射和万古霉素同等浓度的美杀菌素可以有效治疗由金黄色葡萄球菌(Staphylococcus aureus)引起的老鼠全身感染和猫的溃疡病 [35] 。阿肽加定是一种已知的四环羊毛硫抗生素,其分子结构包含19个氨基酸(1890 Da),包括阿肽加定A (Actagardin A) [36] 、脱氧阿肽加定B (Doxy-actagardin B) [37] 和Mchiganin A [38] ,在体外实验和体内的模式动物实验中,均表现出强大的抑制重要的革兰氏阳性病原菌如金黄色葡萄球和化脓性链球菌(Streptococcus pyogenes)活力的能力 [39] 。肉桂霉素家族包括肉桂霉素(Cinnamycin)、耐久霉素(Dramycins)和血管紧张肽转化酶抑制肽(Acovenin),均由放线菌产生,主要用来作为酶抑制剂。
Table 1. Common lantibiotics’ classification, molecular weight and spectrum bactericidal
表1. 常见的羊毛硫抗生素的分类、分子量和杀菌谱
4. 羊毛硫抗生素的合成信号通路及杀菌作用机制
4.1. 羊毛硫抗生素的合成信号通路
羊毛硫抗生素的合成信号通路包括编码羊毛硫抗生素前导肽合成基因lanA,编码负责丝氨酸或苏氨酸脱水的lanB,负责羊毛硫氨酸循环形成的lanC,在某些情况下lanB和lanC可被编码双功能酶的lanM取代 [41] ,并根据是否被取代细分为class I类和class II类,还有编码羊毛硫抗生素调节蛋白的调节基因lanR、lanK,编码转运蛋白基因lanT,编码免疫基因lan-F-E-G,编码用于切除前导肽使之具有生物活性的酶解基因lanP,还有一些其他的基因像lanD负责表皮纤维C端半胱氨酸的氧化脱羧反应,命名为epiD [42] 。在美杀菌素中命名为mrsD [43] (如图3所示)其中mrsFGE为免疫基因,结构合成基因为mrsA,负责后修饰过程的基因mrsD和mrsM,转运基因mrsT和调节基因mrsR1R2K2。合成的具体过程(如图4所示)首先是丝氨酸或者苏氨酸残基在LanB或者LanM的催化下脱氢形成Dha或者Dhb,再在LanC或者LanM的作用下与半胱氨酸的巯基以立体或者局部的方式形成硫醚环。在乳酸链球菌素的合成基因簇中的位点采用nis系列用以标注,编码基因簇片段大小约为14 kb [44] ,包括生物合成、免疫与调控基因,位于乳酸菌染色体基因组上大约70 kb的接合型转座子Tn5031中 [45] ,其生物合成模型展示首先由胞外的刺激信号激活细胞膜上由nisK编码形成的组氨酸激酶并使之磷酸化,磷酸基进一步传递给由nisR编码形成的调控蛋白,再由调控蛋白将前导肽送至修饰蛋白NisB和NisC (分别由nisB和nisC编码合成)完成后修饰环节,与nisT编码的转运蛋白NisT相结合,将前导肽转运出细胞膜,被附在细胞膜外NisP将前导部分切除,从而形成具备生物活性的Nisin大分子 [46] 。同时乳酸菌还具备nisIFEG等相关免疫基因,形成对Nisin的高效耐受性,其中nisI起主效作用,但是必须和nisFEG共同作用才可达到最佳的免疫效果 [47] ,由nisFEG编码的NisFEG可以使胞内的Nisin迅速转移出细胞外,让细胞膜与Nisin结合的机会变小,同时由nisI编码产生的NisI可能位于细胞膜外部作为Nisin的专一性拦截受体并与之结合,避免细胞膜磷脂双分子层与Nisin接触,从而达到对高浓度Nisin的耐受性。

Figure 3. Biosynthetic gene cluster of mersacidine [48]
图3. 美杀菌素(Mersacidine)的生物合成信号通路 [48]
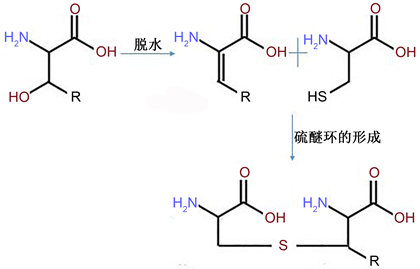
Figure 4. The later modification process of lanthionine [29]
图4. 羊毛硫氨酸的后修饰过程 [29]
4.2. 羊毛硫抗生素的杀菌机制
A类羊毛硫抗生素作用机制分为抑制细胞壁的合成和在细胞膜上打孔两种方式,抑制细胞壁的合成主要是结合在细胞壁合成中起搬运肽聚糖的载体脂质II,从而阻止细胞壁的生物合成导致细菌死亡。在细胞膜上打孔的作用方式是羊毛硫抗生素N端结合脂质II导致其构发生改变 [49] ,C端插入细胞膜上形成孔道 [50] ,导致细胞内大量细胞液(钾离子、谷氨酸和ATP)流出导致细胞的死亡。围绕如何在细胞膜上打孔国内外的学者提出了两种模型:楔子模型和桶板模型 [31] [51] 。在楔子模型中羊毛硫抗生素多肽分子以平行于细胞膜的方向与细胞膜结合,进而细胞膜局部发生扭曲,整个大分子插入细胞膜上的脂质形成孔道。在桶板模型中,羊毛硫抗生素多肽分子中的阳离子通过静电作用去和细胞膜结合,形成一个结合体以垂直方式插入细胞膜上的脂质中,达到膜电位差便可以造成细胞膜出现孔道。B类羊毛硫抗生素也是与细胞壁前体物质脂质II结合致使细胞壁不能正常合成导致死亡,但是结合脂质II的方式与A类不同,是通过静电作用结合的 [52] 。羊毛硫抗生素与细胞膜结合的作用机制分为三步:与细胞膜结合;插入细胞膜;细胞膜形成渗漏(如图5所示)。

Figure 5. Bactericidal mechanism model of lantibiotics [53]
图5. 羊毛硫抗生素的杀菌机制模型 [53]
所有的羊毛硫抗生素必须和靶标菌细胞膜结合 [24] ,羊毛硫抗生素大多带有正电荷,与细胞膜中带有负电荷的磷脂分子结合,革兰氏阳性菌细胞壁较厚,因此对其具有优先的选择性 [54] ,上述介绍的乳酸链球菌素孔道形成即为抗生素大分子N端结合靶标菌细胞膜上的焦磷酸盐 [5] ,C端以平行于细胞膜的方向插入细胞膜形成孔道,但是乳酸链球菌素C端插入经常会跨膜移位,因此乳酸链球菌素插入细胞膜的方向并不固定 [55] 。孔道形成后会发生很快速的毫秒级别的渗漏,孔径是纳米级别的,羊毛硫抗生素与细胞内的阴离子结合并运出细胞外,最后达到一个平衡 [56] 。
5. 羊毛硫抗生素产业化进程与未来展望
目前的研究结果表明,基本上所有的常规抗生素(氨苄青霉素、卡那霉素、壮观霉素等)都或多或少产生了对其具有抗药性的致病株系,因此寻找不易产生抗药性的新型抗生素迫在眉睫。羊毛硫抗生素种类繁多,抑菌谱广且具有较高的活性,不易产生交叉抗性又可使靶标菌不易产生抗性,因此被认为将会在生产中尤其是生物源农药和医学上具有广阔的前景和发展空间。现在的研究虽然已经基本揭示了羊毛硫抗生素的巨大研发潜力,但是全球真正形成商品化的羊毛硫抗生素为数甚少,乳酸链球菌素依旧是最为成功的案例之一。羊毛硫抗生素在农业上的应用主要体现在对已有抗病品种进行基因工程的改造,使改造后的优良品种具有对多种植物病害病原菌的抵抗能力。羊毛硫抗生素在日常生活中主要用作食品防腐剂,例如乳酸链球菌素可以成功的抑制食品中的李斯特菌的感染,具有很高的稳定性和安全性 [57] ,因此也是被美国食品和药物管理局允许使用的食品防腐剂 [58] 。此外,乳酸链球菌素还可以作为保健品用以清新口气 [56] 。与在农业细菌病害和食品防腐剂中发挥作用相比,羊毛硫抗生素在医学上才刚刚起步,有专利报道称由长双歧杆菌产生的羊毛硫抗生素可以有效的对抗引起人体肺炎的沙门氏菌 [59] 。由枯草芽胞杆菌产生的羊毛硫抗生素美杀菌素,是在芽孢杆菌生物防治研究领域研究最为成熟的一种羊毛硫抗生素,可以有效阻止抗药性的葡萄球菌细胞壁的生物合成,在医疗保健上有较大的发展空间 [53] 。
以前从微生物中分离天然存在的羊毛硫抗生素,伴随着现代基因组学和蛋白组学日益成熟并大规模应用于研究的时代,发现并开发新型的羊毛硫抗生素已经取得了巨大的成功,人们不仅可以从细菌基因组中预测新型的羊毛硫抗生素,而且可以改造羊毛硫抗生素的编码基因及化学结构,使之符合现实的需要,在未来几年内人们更可以主动设计新型的羊毛硫抗生素,使得其在农业生产、食品、医学和更多的行业中有更广泛的应用,造福于人类。
基金项目
镇江市农业科学院青年基金(QNJJ2017006)。
文章引用
周华飞,杨红福,束兆林,陈宏州,姚克兵. 细菌羊毛硫抗生素研究进展
Advances in Bacterial Lantibiotics[J]. 微生物前沿, 2019, 08(03): 145-153. https://doi.org/10.12677/AMB.2019.83018
参考文献
- 1. 韩文瑜, 孙长江. 抗菌肽的研究现状与展望[J]. 中国兽药杂志, 2009, 43(10): 11-19.
- 2. 李冠楠, 夏雪娟, 隆耀航, 等. 抗菌肽的研究进展及其应用[J]. 动物营养学报, 2014, 26(1): 17-25.
- 3. Nishie, M., Nagao, J. and Sonomoto, K. (2012) Antibacterial Peptides “Bacteriocins”: An Overview of Their Diverse Characteristics and Applications. Biocontrol Science, 17, 1-16. https://doi.org/10.4265/bio.17.1
- 4. Mcauliffe, O., Ross, R.P. and Hill, C. (2001) Lantibiotics: Structure, Biosynthesis and Mode of Action. FEMS Microbiology Reviews, 25, 285-308. https://doi.org/10.1111/j.1574-6976.2001.tb00579.x
- 5. Willey, J.M. and Wa, V.D.D. (2007) Lantibiotics: Peptides of Diverse Structure and Function. Annual Review of Microbiology, 61, 477-501. https://doi.org/10.1146/annurev.micro.61.080706.093501
- 6. Bierbaum, G. and Sahl, H.G. (2009) Lantibiotics: Mode of Action, Biosynthesis and Bioengineering. Current Pharmaceutical Biotechnology, 10, 2-18. https://doi.org/10.2174/138920109787048616
- 7. Altena, K., Guder, A., Cramer, C. and Bierbaum, G. (2000) Biosynthesis of the Lantibiotic Mersacidin: Organization of a Type B Lantibiotic Gene Cluster. Applied and Environmental Microbiology, 66, 2565-2571. https://doi.org/10.1128/AEM.66.6.2565-2571.2000
- 8. Brötz, H., Bierbaum, G., Reynolds, P.E. and Sahl, H.-G. (2010) The Lantibiotic Mersacidin Inhibits Peptidoglycan Biosynthesis at the Level of Transglycosylation. European Journal of Biochemistry, 246, 193-199. https://doi.org/10.1111/j.1432-1033.1997.t01-1-00193.x
- 9. Allgaier, H., Jung, G., Werner, R.G., Schneider, U. and Zähner, H. (2010) Epidermin: Sequencing of a Heterodetic Tetracyclic 21-Peptide Amide Antibiotic. European Journal of Biochemistry, 160, 9-22. https://doi.org/10.1111/j.1432-1033.1986.tb09933.x
- 10. Hofmeyer, K., Maurel-Zaffran, C., Sink, H., et al. (2014) Production of the Lantibiotic Cinnamycin with Genes Isolated from Streptomyces Cinnamoneus. Clinical Medicine Insights Endocrinology Diabetes, 7, 7-11.
- 11. Kaletta, C., Entian, D.K. and Jung, G. (2010) Prepeptide Sequence of Cinnamycin (Ro 09-0198): The First Structural gene of a Duramycin-Type Lantibiotic. European Journal of Biochemistry, 199, 411-415. https://doi.org/10.1111/j.1432-1033.1991.tb16138.x
- 12. García-Cayuela, T., Requena, T., Martínez-Cuesta, M.C. and Peláez, C. (2017) Rapid Detection of Lactococcus lactis Isolates Producing the Lantibiotics Nisin, Lacticin 481 and Lacticin 3147 Using MALDI-TOF MS. Journal of Microbiological Methods, 139, 138-142. https://doi.org/10.1016/j.mimet.2017.06.002
- 13. Mills, S., Griffin, C., O’Connor, P.M., et al. (2017) A Mul-ti-Bacteriocin Cheese Starter System Comprising Nisin and Lacticin 3147 in Lactococcus lactis, in Combination with Plantaricin from Lactobacillus plantarum. Applied & Environmental Microbiology, 83, 717-799. https://doi.org/10.1128/AEM.00799-17
- 14. Weil, H.P., Beck-Sickinger, A.G., Metzger, J., et al. (2010) Biosynthesis of the Lantibiotic Pep5. Isolation and Characterization of a Prepeptide Containing Dehydroamino Acids. European Journal of Biochemistry, 194, 217-223. https://doi.org/10.1111/j.1432-1033.1990.tb19446.x
- 15. Castiglione, F., Lazzarini, A., Carrano, L., et al. (2008) Determining the Structure and Mode of Action of Microbisporicin, a Potent Lantibiotic Active against Multiresistant Pathogens. Chemistry Biology, 15, 22-31. https://doi.org/10.1016/j.chembiol.2007.11.009
- 16. Foulston, L.C. and Bibb, M.J. (2010) Microbisporicin Gene Cluster Reveals Unusual Features of Lantibiotic Biosynthesis in Actinomycetes. Proceedings of the National Academy of Sciences of the United States of America, 107, 13461-13466. https://doi.org/10.1073/pnas.1008285107
- 17. Foulston, L. and Bibb, M. (2011) Feed-Forward Regulation of Microbisporicin Biosynthesis in Microbispora corallina. Journal of Bacteriology, 193, 3064. https://doi.org/10.1128/JB.00250-11
- 18. 刘宝生, 张锦华, 罗军荣. 细菌素及其应用的研究进展[J]. 中国畜牧兽医, 2013, 40(5): 123-128.
- 19. Kuipers, O.P., Beerthuyzen, M.M., Siezen, R.J. and De Vos, W.M. (2010) Characterization of the Nisin Gene Cluster nisABTCIPR of Lactococcus lactis. Requirement of Expression of the nisA and nisI genes for Development of Immunity. European Journal of Biochemistry, 216, 281-291. https://doi.org/10.1111/j.1432-1033.1993.tb18143.x
- 20. Mulders, J.W.M., Boerrigter, I.J., Rollema, H.S., Siezen, R.J. and De Vos, W.M. (2010) Identification and Characterization of the Lantibiotic Nisin Z, a Natural Nisin Variant. European Journal of Biochemistry, 201, 581-584. https://doi.org/10.1111/j.1432-1033.1991.tb16317.x
- 21. Buchman, G.W., Banerjee, S. and Hansen, J.N. (1988) Structure, Expression, and Evolution of a Gene Encoding the Precursor of Nisin, a Small Protein Antibiotic. Journal of Biological Chemistry, 263, 16260.
- 22. Kaletta, C. and Entian, K.D. (1989) Nisin, a Peptide Antibiotic: Cloning and Sequencing of the nisA Gene and Posttranslational Processing of Its Peptide Product. Journal of Bacteriology, 171, 1597-1601. https://doi.org/10.1128/jb.171.3.1597-1601.1989
- 23. Schnell, N., Entian, K.D., Schneider, U., et al. (1988) Prepeptide Sequence of Epidermin, a Ribosomally Synthesized Antibiotic with Four Sulphide-Rings. Nature, 333, 276-278. https://doi.org/10.1038/333276a0
- 24. Todorov, S.D. (2009) Bacteriocins from Lactobacillus plantarum-Production, Genetic Organization and Mode of Action: Producao, organizacao genetica e modo de acao. Brazilian Journal of Microbiology, 40, 209-221. https://doi.org/10.1590/S1517-83822009000200001
- 25. 田文利, 吴琼. 乳酸链球菌素(Nisin)的研究进展[J]. 食品工业, 2000, 21(3): 28-30.
- 26. 吕淑霞, 白泽朴, 代义, 等. 乳酸链球菌素(Nisin)抑菌作用及其抑菌机理的研究[J]. 中国酿造, 2008, 27(9): 87-91.
- 27. Slijper, M., Hilbers, C.W., Konings, R.N.H. and van de Ven, F.J.M. (1989) NMR Studies of Lantibiotics Assignment of the 1H-NMR Spectrum of Nisin and Identification of Interresidual Contacts. FEBS Letters, 252, 22-28.
- 28. Bauer, R. and Dicks, L.M.T. (2005) Mode of Action of Lipid II-Targeting Lantibiotics. International Journal of Food Microbiology, 101, 201-216. https://doi.org/10.1016/j.ijfoodmicro.2004.11.007
- 29. Cortes, J. (2014) Lantibiotics and Similar Peptides Produced by and Active on Gram-Positives: Discovery, Development and Perspectives. In: Marinelli, F. and Genilloud, O., Eds., Antimicrobials, Springer, Berlin, Heidelberg, 141-158.
- 30. 顾觉奋, 何文杰. Lantibiotic: 一类新颖的抗菌肽研究进展[J]. 国外医药抗生素分册, 2001, 22(6): 274-278.
- 31. Asaduzzaman, S.M. and Sonomoto, K. (2009) Lantibiotics: Diverse Activities and Unique Modes of Action. Journal of Bioscience & Bioengineering, 107, 475-487. https://doi.org/10.1016/j.jbiosc.2009.01.003
- 32. Imke, W., Tim, B.T., Bonelli, R.R., et al. (2010) The Mode of Action of the Lantibiotic Lacticin 3147-a Complex Mechanism Involving Specific Interaction of Two Peptides and the Cell Wall Precursor Lipid II. Molecular Microbiology, 61, 285-296. https://doi.org/10.1111/j.1365-2958.2006.05223.x
- 33. Gross, E. and Kiltz, H.H. (1973) The Number and Nature of α, β-Unsaturated Amino Acids in Subtilin. Biochemical and Biophysical Research Communications, 50, 559-565.
- 34. Gross, E. and Morell, J.L. (1967) The Presence of Dehydroalanine in the Antibiotic Nisin and Its Relationship to Activity. Journal of the American Chemical Society, 89, 2791-2792. https://doi.org/10.1021/ja00987a084
- 35. Chatterjee, S., Chatterjee, S., Lad, S.J., et al. (1992) Mersacidin, a New Antibiotic from Bacillus. Fermentation, Isolation, Purification and Chemical Characterization. Journal of Antibiotics, 45, 832-838.
- 36. Zimmermann, N., Metzger, J.R.W. and Jung, G. (1995) The Tetracyclic Lantibiotic Actagardine 1H-NMR and 13C-NMR Assignments and Revised Primary Structure. European Journal of Biochemistry, 228, 786-797.
- 37. Steven, B., Appleyard, A.N., Jesús, C. and Dawson, M.J. (2010) Organization of the Biosynthetic Genes Encoding Deoxyactagardine B (DAB), a New Lantibiotic Produced by Actinoplanes liguriae NCIMB41362. Journal of Antibiotics, 63, 351-358. https://doi.org/10.1038/ja.2010.48
- 38. Holtsmark, I., Mantzilas, D., Eijsink, V.G.H. and Brurberg, M.B. (2006) Purification, Characterization, and Gene Sequence of Michiganin A, an Actagardine-Like Lantibiotic Produced by the Tomato Pathogen Clavibacter michiganensis subsp. michiganensis. Applied and En-vironmental Microbiology, 72, 5814-5821. https://doi.org/10.1128/AEM.00639-06
- 39. Zimmermann, N., Metzger, J.W. and Jung, G. (2010) The Tetracyclic Lantibiotic Actagardine 1H-NMR and 13C-NMR Assignments and Revised Primary Structure. The FEBS Journal, 228, 786-797.
- 40. 姚惠源, 张晖, 管骁, 等. 一种燕麦蛋白ACE抑制肽的制备方法[P]. 中国专利, CN1944663. 2007-04-11.
- 41. Lili, X., Miller, L.M., Champak, C., et al. (2004) Lacticin 481: In Vitro Reconstitution of Lantibiotic Synthetase Activity. Science, 303, 679-681.
- 42. Kupke, T., Stevanovi, S., Sahl, H.G. and Götz, F. (1992) Purification and Characterization of EpiD, a Flavoprotein Involved in the Biosynthesis of the Lantibiotic Epidermin. Journal of Bacteriology, 174, 5354-5361. https://doi.org/10.1128/jb.174.16.5354-5361.1992
- 43. Florian, M., Schmid, D.G., Karsten, A., Bierbaum, G. and Kupke, T. (2002) The Flavoprotein MrsD Catalyzes the Oxidative Decarboxylation Reaction Involved in Formation of the Peptidoglycan Biosynthesis Inhibitor Mersacidin. Journal of Bacteriology, 184, 1234-1243. https://doi.org/10.1128/JB.184.5.1234-1243.2002
- 44. 贾士芳, 还连栋, 王连琴. Nisin生物合成有关基因的遗传分析[J]. 中国生物工程杂志, 1997, 17(4): 35-39.
- 45. Horn, N., Swindell, S., Dodd, H. and Gasson, M. (1991) Nisin Biosynthesis Genes Are Encoded by a Novel Conjugative Transposon. Molecular and General Genetics MGG, 228, 129-135. https://doi.org/10.1007/BF00282457
- 46. Entian, K.D. and de Vos, W.M. (1996) Genetics of Subtilin and Nisin Biosyntheses: Biosynthesis of Lantibiotics. Antonie Van Leeuwenhoek, 69, 109-117. https://doi.org/10.1007/BF00399416
- 47. Torsten, S., Stefan, H., Irina, S. and Entian, K.-D. (2003) Function of Lactococcus lactis Nisin Immunity Genes nisI and nisFEG after Coordinated Expression in the Surrogate Host Bacillus subtilis. Journal of Biological Chemistry, 278, 89. https://doi.org/10.1074/jbc.M207237200
- 48. Anna Maria, H., Jasmin, D., Christiane, S., et al. (2011) Expression of the Lantibiotic Mersacidin in Bacillus amyloliquefaciens FZB42. PLoS ONE, 6, e22389. https://doi.org/10.1371/journal.pone.0022389
- 49. Wiedemann, I., Breukink, E., Kraaij, C.V., et al. (2001) Specific Binding of Nisin to the Peptidoglycan Precursor Lipid II Combines Pore Formation and Inhibition of Cell Wall Biosynthesis for Potent Antibiotic Activity. Journal of Biological Chemistry, 276, 1772-1779. https://doi.org/10.1074/jbc.M006770200
- 50. Shang-Te, H., Eefjan, B., Ben, D.K., et al. (2002) Mapping the Targeted Membrane Pore Formation Mechanism by Solution NMR: The Nisin Z and Lipid II Interaction in SDS Micelles. Biochemistry, 41, 7670-7676. https://doi.org/10.1021/bi025679t
- 51. Park, S.C., Park, Y. and Hahm, K.S. (2011) The Role of Antimicrobial Peptides in Preventing Multidrug-Resistant Bacterial Infections and Biofilm Formation. International Journal of Molecular Sciences, 12, 5971-5992. https://doi.org/10.3390/ijms12095971
- 52. Siragusa, G.R., Cutter, C.N. and Willett, J.L. (1999) Incorporation of Bacteriocin in Plastic Retains Activity and Inhibits Surface Growth of Bacteria on Meat. Food Microbiology, 16, 229-235. https://doi.org/10.1006/fmic.1998.0239
- 53. Dischinger, J., Chipalu, S.B. and Bierbaum, G. (2014) Lantibiotics: Promising Candidates for Future Applications in Health Care. International Journal of Medical Microbiology, 304, 51-62. https://doi.org/10.1016/j.ijmm.2013.09.003
- 54. Fang, T.J. and Tsai, H.C. (2003) Growth Patterns of Escherichia coli O157:H7 in Ground Beef Treated with Nisin, Chelators, Organic Acids and Their Combinations Immobilized in Calcium Alginate Gels. Food Microbiology, 20, 243-253. https://doi.org/10.1016/S0740-0020(02)00081-3
- 55. Sahl, H.G., Kordel, M. and Benz, R. (1987) Voltage-Dependent Depolarization of Bacterial Membranes and Artificial Lipid Bilayers by the Peptide Antibiotic Nisin. Archives of Microbiology, 149, 120-124. https://doi.org/10.1007/BF00425076
- 56. Kraaij, C.V., Vos, W.M.D., Siezen, R.J. and Kuipers, O.P. (1999) Lantibiotics: Biosynthesis, Mode of Action and Applications. Natural Product Reports, 16, 575. https://doi.org/10.1039/a804531c
- 57. Duhan, J.S., Nehra, K., Gahlawat, S.K., Saharan, P. and Surekha, D. (2013) Bacteriocins from Lactic Acid Bacteria. In: Salar, R., Gahlawat, S., Siwach, P. and Duhan, J., Eds., Biotechnology: Prospects and Applications, Springer, New Delhi, 127-141. https://doi.org/10.1007/978-81-322-1683-4_11
- 58. 孙健, 郑珩, 徐寒梅. 羊毛硫抗生素的研究进展及其应用[J]. 国外医药抗生素分册, 2013, 34(2): 49-52.
- 59. O’Sullivan, D.J. and Lee, J.H. (2015) Lantibiotics and Uses Thereof.
NOTES
*通讯作者。
