Advances in Clinical Medicine
Vol.
11
No.
06
(
2021
), Article ID:
43612
,
11
pages
10.12677/ACM.2021.116418
肝细胞癌肿瘤突变负荷与肿瘤微环境免疫浸润细胞关联性研究
曲家麟,王力,姜曼,张晓春*
青岛大学附属医院肿瘤精准医学中心,山东 青岛
收稿日期:2021年5月28日;录用日期:2021年6月19日;发布日期:2021年6月30日

摘要
目的:肝细胞癌是我国常见肿瘤之一,并占癌症死亡人数的第二位。随着免疫治疗的普及,免疫检查点抑制剂也逐步应用于肝癌,部分患者取得了良好的治疗效果。但是目前仍缺少准确地预测免疫检查点抑制剂的生物标志物。肿瘤突变负荷及免疫浸润细胞是常用的免疫检查点抑制剂的生物标志物,可以有效预测其疗效,在恶性黑色素瘤及非小细胞肺癌中已经得到证实。但在肝癌中此类研究尚少,为了探究肿瘤突变负荷及免疫浸润细胞之间是否存在协同关系以便更好预测免疫检查点抑制剂在肝癌中的疗效,我们展开了本次研究。方法:从TCGA数据库中下载肝癌患者的基因表达数据、基因突变数据及临床数据。将所有患者根据肿瘤突变负荷分为高突变负荷组及低突变负荷组,从两组患者的数据中鉴定出差异表达最明显的基因。利用功能富集分析及蛋白质–蛋白质相互作用网络分析差异基因的生物学功能。CIBERSORT算法用于分析两组患者之间肿瘤微环境免疫浸润细胞的特征,Kaplan-Meier生存分析用以分析两组之间患者的生存时间有无差别。结果:TP53是肝细胞癌中突变频率最高的基因,在高肿瘤突变负荷组及低肿瘤突变复合组中,MAGEA3、MAGEA6及MAGEA12基因表达差异最为显著,其功能与细胞增殖、上皮–间质转化等恶性表型密切相关。在肿瘤微环境免疫浸润细胞差别中,低肿瘤突变复合组的记忆性CD4+ T细胞数量显著高于高肿瘤突变复合组,且生存期明显延长。结论:在肝细胞癌患者中,低肿瘤突变负荷组的CD4+ T细胞数量显著高于高肿瘤突变复合组,并且有更好的临床预后。这显示在肝细胞癌中,以TMB高低作为判断肝细胞癌免疫检查点抑制剂的唯一生物标志物尚有待商榷,应结合更多指标综合评估。
关键词
肝细胞癌,免疫检查点抑制剂,肿瘤突变负荷,免疫浸润细胞

Identification of the Relationships between Tumor Mutation Burden with Immune Infiltrates in Liver Hepatocellular Carcinoma
Jialin Qu, Li Wang, Man Jiang, Xiaochun Zhang*
Cancer Precision Medicine Center, The Affiliated Hospital of Qingdao University, Qingdao Shandong
Received: May 28th, 2021; accepted: Jun. 19th, 2021; published: Jun. 30th, 2021

ABSTRACT
Background: Hepatocellular carcinoma (HCC) is one of the most common tumors in China, and accounts for the second largest number of cancer deaths. With the popularity of immunotherapy, immune checkpoint inhibitors are gradually applied to liver cancer, and some patients have achieved good therapeutic effects. However, there is still a lack of accurate biomarkers to predict immune checkpoint inhibitors. Tumor mutation load and immune infiltrating cells are commonly used as biomarkers of immune checkpoint inhibitors, which can effectively predict efficacy. It has been confirmed in malignant melanoma and non-small cell lung cancer. In order to explore whether there is a synergistic relationship between tumor mutation load and immune infiltrating cells, so as to better predict the efficacy of immune checkpoint inhibitors in liver cancer, we launched this study. Methods: The mutation data, clinical data and gene expression data of liver hepatocellular carcinoma patients were downloaded from TCGA database. Differentially expressed genes were identified from high and low TMB groups. Functional enrichment analyses and protein-protein interaction (PPI) network analysis were used to identify the functions and pathway of the DEGs. And immune cell infiltration signatures were evaluated by CIBERSORT algorithm. Results: TP53 is the gene with the highest mutation frequency in hepatocellular carcinoma. The expression of magea3, magea6 and magea12 genes is the most significant in high tumor mutation load group and low tumor mutation complex group. Its function is closely related to cell proliferation, epithelial mesenchymal transition and other malignant phenotypes. The number of memory CD4+ T cells in low mutation group was significantly higher than that in high mutation group, and the survival time was significantly longer. Conclusion: In HCC patients, the number of CD4+ T cells in low mutation load group was significantly higher than that in high mutation complex group, and the clinical prognosis was better. This suggests that high mutation load may not be a positive prognostic marker of immunosuppressive agents in HCC.
Keywords:Hepatocellular Carcinoma, Immune Checkpoint Inhibitors, Tumor Mutation Burden, Immune Infiltrating Cells

Copyright © 2021 by author(s) and Hans Publishers Inc.
This work is licensed under the Creative Commons Attribution International License (CC BY 4.0).
http://creativecommons.org/licenses/by/4.0/


1. 概论
肝细胞癌是全世界发病率第五位及死亡率第二位的恶性肿瘤。我国肝癌病例发病数占全世界的50%左右,其死亡率仅次于肺癌 [1] [2]。手术切除或肝移植是治疗肝癌最有效的方法,但由于起病隐匿,大部分患者就诊时已处于晚期,丧失手术机会。对于无法手术的晚期肝癌患者,以铂类为基础的静脉化疗方案仍是一线用药。尽管随着索拉非尼、瑞戈非尼等小分子酪氨酸激酶抑制剂先后投入临床,改善了肝癌患者的生存期,但总体而言,肝癌患者的五年生存率仍然不甚理想 [3] [4]。随着免疫学的进展,肿瘤细胞发生免疫逃逸的关键因素——免疫检查点被发现,以免疫检查点抑制剂为代表的免疫治疗成为肿瘤治疗的新近突破性进展,在包括恶性黑色素瘤、非小细胞肺癌等肿瘤的治疗中取得了令人振奋的效果。多种靶向CTLA-4、PD-1等免疫检查点的单克隆抗体包括伊匹木单抗、帕博利珠单抗及纳武利尤单抗等免疫检查点抑制剂被批准用于多种肿瘤的一线或多线治疗 [5]。其中,PD-1帕博利珠单抗已在全球获批用于肝细胞癌的二线治疗。除此之外,美国FDA已授予靶向抗癌药仑伐替尼与抑制剂帕博利珠单抗组合疗法突破性药物资格,用于一线治疗不适合局部区域治疗的晚期不可切除性肝细胞癌患者,揭开了肝癌治疗新的序幕 [6] [7]。
然而,免疫检查点抑制剂的总体有效率仅为30% [8]。并且由于免疫检查点抑制剂费用昂贵,因此目前迫切需要能够准确预测疗效的生物标志物以筛选最佳适应症的患者,尽可能实现精准医疗并节省经济资源。免疫浸润细胞特别是T细胞是机体抗肿瘤免疫反应的效应器,如肿瘤微环境缺乏足够的免疫细胞浸润则患者对免疫检查点抑制剂无反应,先前研究证实免疫浸润细胞与免疫检查点抑制剂的临床治疗疗效密切相关 [9]。与此同时,越来越多的研究证实肿瘤突变负荷(tumor mutation burden, TMB)与多种癌症类型免疫检查点抑制剂治疗后较长的生存期有关,提示TMB可以作为一种免疫检查点抑制剂治疗疗效的预测生物标志物 [10]。肿瘤突变负荷定义为每百万碱基中被检测出的体细胞基因编码错误、碱基替换、基因插入或缺失错误的总数。这些肿瘤细胞的突变可以被转录和翻译,形成新的肿瘤抗原,成为免疫细胞识别的标记和靶点 [11]。肿瘤突变负荷越多,形成的新抗原就越多,肿瘤的免疫原性越强,这能够增强免疫治疗的疗效 [12]。
在肝癌中,肿瘤突变负荷与肿瘤浸润细胞这两种影响免疫治疗疗效的两种主要影响因素之间是否存在协同作用,高肿瘤突变负荷能否募集更多肿瘤浸润细胞至肿瘤微环境中以增强免疫检查点抑制剂的疗效,这些问题在一定程度上能够解释肿瘤突变负荷是否可以作为肝癌免疫治疗的生物标志物。因此,在这篇文章中,我们探讨了肝细胞癌中肿瘤突变负荷与肿瘤浸润细胞之间的关系,这项研究的发现可能为肝细胞癌患者中TMB作为预测疗效的生物标志物的确定提供部分依据。
2. 方法
1) 数据预处理
从肿瘤基因图谱(The Cancer Genome Atlas, TCGA)数据库中下载美国和日本肝细胞癌患者的体细胞突变数据。利用maftools软件包对突变数据进行分析和总结。通过肝癌特异性突变基因计算得到每个患者的TMB值。limma软件包用以分析基因表达数据。调整后的P值(adj. P) < 0.05和|logFC| ≥ 1.0作为确定差异基因表达的截止值。pheatmap包制作热图以可视化数据。
2) 基因功能和通路富集分析
基因本体(The Gene Ontology, GO)数据库主要包括三类:生物过程(Biological Process, BP)、细胞成分(Cellular Component, CC)和分子功能(Molecular Function, MF) [13]。京都基因和基因组百科全书(The Kyoto Encyclopedia of Genes and Genomes, KEGG)数据库收集基因组、化学和系统功能信息 [14]。使用clusterProfiler软件包进行基因本体富集分析和京都基因与基因组百科全书途径富集分析,富集阈值为P < 0.05 [15]。
3) 建立蛋白互作网络
STRING数据库和Cytoscape用于构建和分析蛋白质–蛋白质相互作用网络 [16] [17]。截止标准设定为综合得分 > 0.4。Cytoscape插件CytoHubba通过从12种拓扑分析方法中找到前30个基因的交叉点来识别hub基因,并从每种方法的结果中选出前10个hub基因 [18]。
4) 鉴定肿瘤浸润性免疫细胞
CIBERSORT是一种分析工具,利用基因表达数据估计混合细胞群中的特定细胞类型。该算法由1000个排列和LM22签名运行 [19]。用CIBERSORT法定量肝细胞癌标本肿瘤浸润性免疫细胞的类型和数量。
5) TMB分组患者的生存分析
以突变频率中位数为阈值,将肝癌患者分为高TMB组及低TMB组。通过logrank检验获得Kaplan-Meier生存曲线。采用单因素方差分析、Tukey多重比较事后检验和非配对双尾t检验分析临床特征与TMB的相关性。
3. 结论
1) 肝细胞癌患者中基因突变类型的综合分析
在本次研究中,从TCGA数据库中下载肝细胞癌患者基因突变的数据。使用maftools软件包对突变
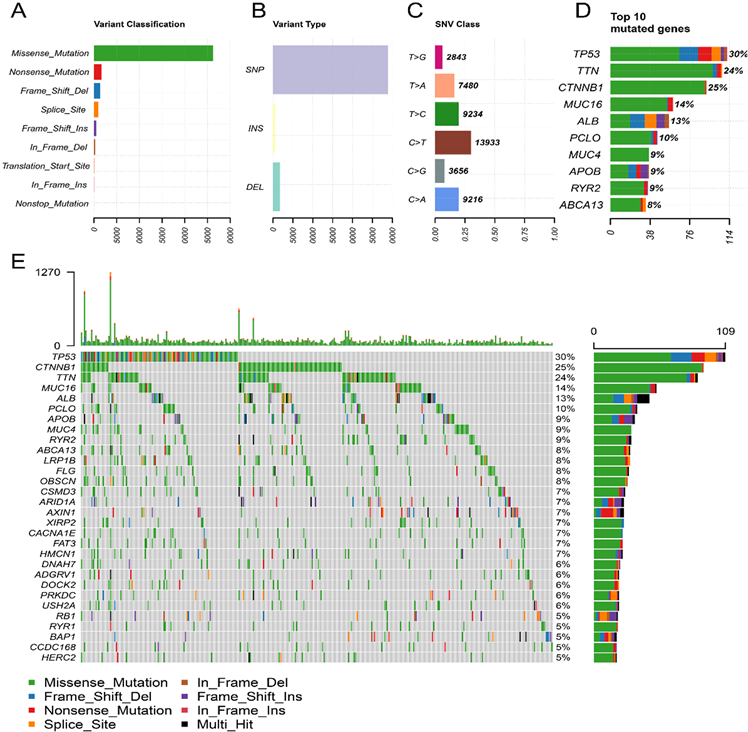
Figure 1. (A) (B) Primary genetic alterations in liver hepatocellular carcinoma; (C) Variant classification and type of genetic alterations; (D) (E) The SNV class of liver hepatocellular carcinoma
图1. (A) (B) 原发性肝细胞癌的遗传突变;(C) 遗传突变的类型;(D) (E) 肝细胞癌的单核苷酸变异类型及最常见的10种基因突变
数据进行分析和总结。根据变异效应预测因子对突变进行进一步分类。结果显示,肝细胞癌患者最常见的突变是错义突变(图1(A))。单核苷酸多态性(single nucleotide polymorphism, SNP)是最常见的突变类型(图1(B))。肝细胞癌中最常见的SNP类型是T>G颠换(图1(C))。TP53 (30%)、CTNBB1 (25%)、TTN (24%)、MUC16 (14%)、ALB (13%)、PCLO (10%)、APOB (9%)、MUC4 (9%)、RYR2 (9%)、ABCA13 (8%)是肝细胞癌中最常见的10个突变基因(图1(D)、图1 (E))。
2) 肿瘤突变负荷与肝细胞癌患者的预后关系
TMB的计算方法是非同义蛋白编码变异数除以总的基因组长度。在我们的研究中,我们根据TMB中位数将所有肝癌患者分为低TMB组和高TMB组。Kaplan-Meier生存分析显示,在男性患者亚组中,低TMB患者的生存率高于高TMB患者(图2(A))。但在女性患者亚组中,TMB与患者的生存时间没有明显关系(图2(B))。接下来,我们分析了肿瘤分期和TMB之间的关系,结果显示TMB与临床分期无关(图2(C))。然而,男性肝癌患者的TMB水平明显较女性患者为高(图2(D))。
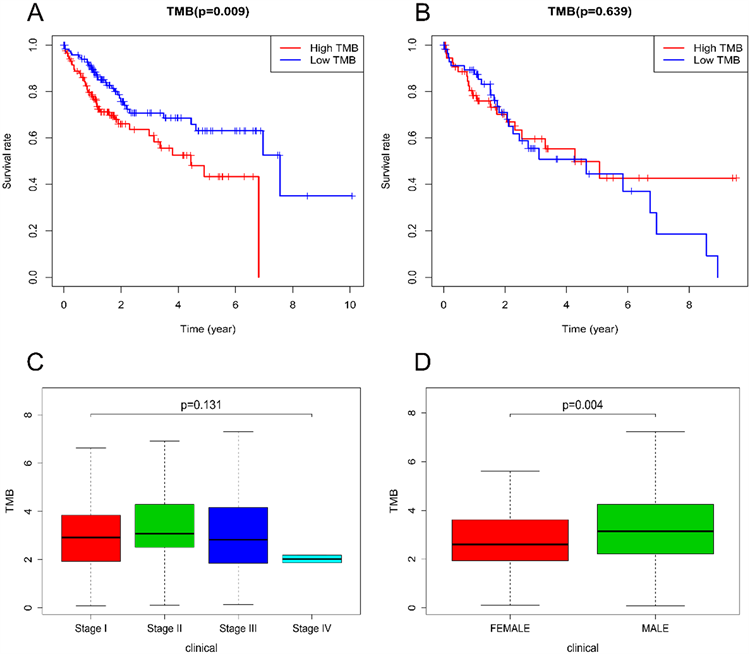
Figure 2. Correlation of TMB with prognosis, gender and tumor grades of LIHC patients. (A) Kaplan-Meier survival curve of male patients; (B) Kaplan-Meier survival curve of female patients; (C) The TMB showed no statistically significant differences at different pathological stages; (D) Higher TMB level correlated with gender
图2. 与原发性肝细胞癌患者预后、性别及肿瘤分级之间的关系。(A) 男性患者的K-M生存曲线;(B) 女性患者的K-M生存曲线;(C) TMB在不同肿瘤分期中无明显差异;(D) TMB水平与性别相关
3) 不同TMB分组之间的基因表达差异
在本项研究中,我们分析并鉴定了高MB组与低MB组肝癌患者之间差异表达的基因。两组之间共有198个基因差异表达,其中高TMB组患者有170个基因表达量较低TMB组明显上调,28个基因明显下调。这些基因通过热图进行可视化处理(图3)。

Figure 3. Comparison of the gene expression profiles in different TMB groups. The heatmap plot show the 198 DEGs (170 downregulated and 28 upregulated) identified based on the TMB
图3. 不同TMB组基因表达差异。热图显示了高TMB组和低TMB组差异表达最明显的198个基因(其中170个下调,28个上调)
4) 差异基因功能分析
通过GO富集分析确定在TMB高表达组及低表达组之间表达差异最明显的198个基因的生物学功能。分析结果显示,这些差异基因主要与细胞外基质组织、细胞外结构组织、细胞粘附的负调控和结缔组织的发育等功能有关有关。此外,细胞成分分析显示这些差异基因主要与与细胞外基质、胶原的形成有关。此外,这些基因表达的蛋白功能主要是细胞外基质结构成分、糖胺聚糖结合和硫化合物结合(图4(A))。我们还进行了KEGG途径富集分析,分析结果显示这些差异基因主要参与细胞间粘附、上皮–间质转化(Epithelial-Mesenchymal Transition, EMT)等信号通路,这些通路与恶性肿瘤的侵袭及转移密切相关。这在一定程度上解释了TMB低表达组患者的预后好于TMB高表达组患者的原因(图4(B))。
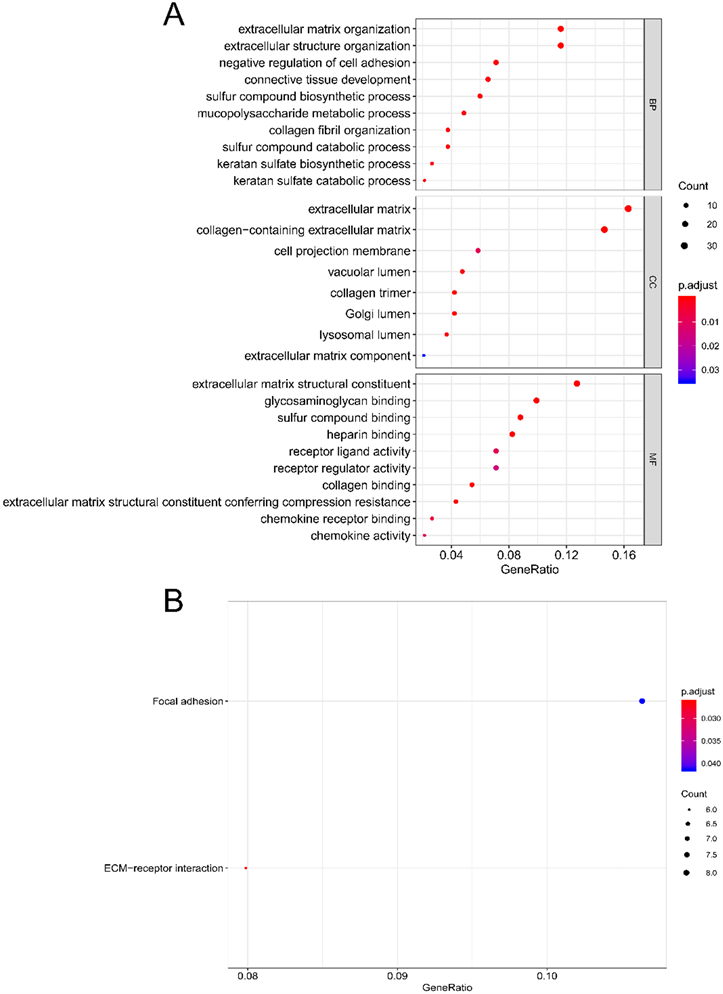
Figure 4. (A) Functional and pathway enrichment analysis; (B) Enrichment of biological process, enrichment of cellular component, and enrichment of molecular function
图4. (A) 功能及通路富集分析;(B) 生物学功能、细胞组分及分子功能分析
5) 蛋白互作网络分析
差异基因–蛋白互作网络由198个节点和295个边缘组成,包括28个上调基因和170个下调基因(图5(A))。通过CytoHubba分析和探索蛋白互作网络的重要节点和子网络。前10位的hub基因分别为COL1A1、VCAN、POSTN、LUM、DCN、FMOD、MAGEA1、MAGEA12、MAGEA3和MAGEA6。通过使用MCODE从网络获得重要模块,包括8个节点和27个边缘(图5(B))。此外,我们发现MAGEA12、MAGEA3和MAGEA6参与了GO及KEGG分析。
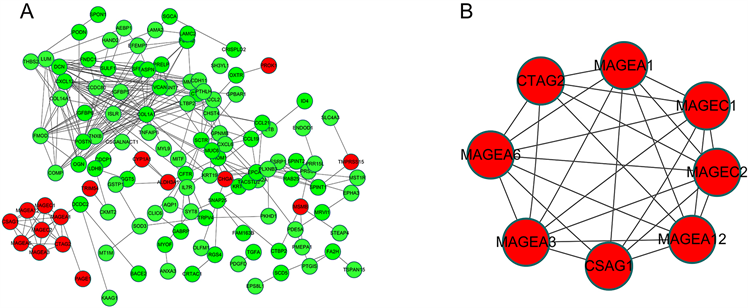
Figure 5. (A) PPI network and module analysis; (B) Protein-protein interaction networks of DEGs; significant module selected from PPI network. red nodes, upregulated genes; yellow nodes, downregulated genes
图5. (A) PPI网络和模块分析;(B) 蛋白质–差异表达基因的蛋白质相互作用网络;PPI中选择的重要模块。红色节点:上调基因;黄色节点,下调基因
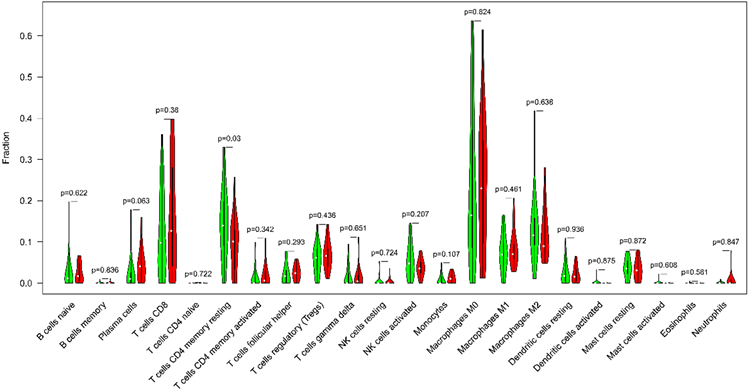
Figure 6. Correlation of TMB with immune signatures in liver hepatocellular carcinoma
图6. 肝细胞癌中TMB与免疫浸润细胞类型的相关性
6) 肿瘤浸润免疫细胞分析
在此项研究中,CIBERSORT算法用以评估了TMB与肝癌微环境中肿瘤浸润免疫细胞的关系。以P < 0.05为阈值,CIBERSORT产生的免疫细胞群的推断分数的结果被认为是准确的。最后,分析结果显示低TMB组肿瘤微环境中CD4+记忆性T淋巴细胞显著高于高TMB组(图6)。
4. 讨论
肿瘤是一种基因性疾病,由DNA中的体细胞突变累积引起 [20]。在不同的肿瘤类型和个体肿瘤中,每种类型基因改变的频率存在显著差异 [21]。基因突变增加了肿瘤的免疫原性。目前,肝细胞癌的总生存率(OS)随着免疫检查点抑制剂投入临床应用而得以延长 [1]。TMB正在成为一种潜在的生物标记物,用于预测免疫检查点阻断疗法对黑色素瘤、肺癌、膀胱癌和头颈部鳞癌癌的疗效 [20] [22]。
TMB在几乎所有类型的癌症中均存在,且TMB在同一癌症类型中也有显著差异 [23]。TMB反映了肿瘤基因突变所产生的肿瘤相关新抗原的存在,从而增加了肿瘤微环境中淋巴细胞的浸润 [24] [25]。肿瘤微环境包括免疫细胞、内皮细胞、成纤维细胞、间充质细胞和细胞外成分,这些细胞外成分围绕着肿瘤细胞,并由血管网络滋养 [26]。一些研究表明,肿瘤微环境不仅在肿瘤的发生、发展和转移过程中起着关键作用,而且对治疗效果有着深远的影响 [27] [28]。
在本研究中,我们发现TMB能够影响男性肝癌患者的预后并且男性患者的TMB明显高于女性患者。然后,我们在不同的TMB亚组之间鉴定了198个差异表达基因。其中许多基因涉及免疫应答和趋化因子信号通路。功能和途径富集分析表明,这些差异基因影响细胞外基质组织、细胞外基质、细胞外基质结构成分、细胞粘附和上皮–间质转化。这些结果提示,TMB与肿瘤微环境密切相关,这些TMB相关基因可能引起肿瘤微环境的改变。PPI网络和模块分析表明,MAGEA12、MAGEA3和MAGEA6是所有DEG中的重要枢纽基因。
黑色素瘤相关抗原A (melanoma-associated antigen-A, MAGE-A)家族是一个由12个同源基因(MAGE-A1到-A12)组成的多基因家族 [29]。据报道,表达MAGE-A的癌细胞可以增强免疫反应 [30] [31] [32]。一项研究表明,上尿路癌细胞中MAGE-A的表达与CD3、CD8和CD45RO阳性T淋巴细胞的浸润有关,而与CD20阳性B淋巴细胞的浸润无关 [33]。此外,在肝癌和肺癌的外周血中检测到MAGEA反应性CD8阳性T淋巴细胞 [34] [35]。因此,我们的结果提示MAGEA12、MAGEA3和MAGEA6可能在肝细胞癌的肿瘤微环境及局部抗肿瘤免疫应答调控中起重要作用。
免疫细胞浸润是肿瘤微环境的标志之一。肿瘤微环境有一个复杂的趋化因子网络,影响浸润细胞的数量和表型 [36]。在我们的研究中,我们发现低TMB组有较高的记忆性CD4+ T细胞。这一结果表明,TMB可影响免疫细胞的浸润特征。记忆性CD4+ T细胞能够长时间记忆肿瘤细胞的免疫特征,是免疫治疗有效后长时间肿瘤消退的原因之一 [37]。CD4+ T细胞可以通过多种方式靶向肿瘤细胞,既可以通过细胞溶解机制直接清除肿瘤细胞,也可以通过调节肿瘤微环境间接靶向肿瘤细胞 [38] [39]。此外,在次级淋巴器官中,CD4+ T细胞增加了B细胞和细胞毒性T淋巴细胞反应的数量和质量 [40] [41]。CD4+ T细胞还可以帮助启动CD8+ T细胞的活化程序,通过多种分子机制增强细胞毒性T淋巴细胞的反应,使细胞毒性T淋巴细胞获得更强的杀瘤活性 [42]。结果表明,TMB与免疫微环境密切相关,低TMB组中高表达的CD4+ T细胞可能抑制肿瘤的发生和转移,增加免疫治疗的应答率。
当然,我们的研究也存在一些不可避免的局限性:首先,我们的结果没有考虑年龄、肿瘤组织学类型等特征进行详细的分类。其次,我们的研究结果没有得到临床研究或实验的证实。因此,我们期待更多的临床研究来证实我们的结果。并且有必要进行进一步的分子生物学来证实我们的研究结果。
5. 结论
在本研究中,AGEA12、MAGEA3和MAGEA6可能在肝细胞癌的肿瘤微环境免疫调节中起关键作用。低TMB男性肝癌患者预后较好。此外,低TMB组与更多的记忆性CD4+ T细胞数量显著相关。我们希望本研究能为确定肝细胞癌免疫治疗的有效生物标志物提供一个新的思路。
基金项目
山东省自然科学基金(ZR2018MH022)。
文章引用
曲家麟,王 力,姜 曼,张晓春. 肝细胞癌肿瘤突变负荷与肿瘤微环境免疫浸润细胞关联性研究
Identification of the Relationships between Tumor Mutation Burden with Immune Infiltrates in Liver Hepatocellular Carcinoma[J]. 临床医学进展, 2021, 11(06): 2880-2890. https://doi.org/10.12677/ACM.2021.116418
参考文献
- 1. Nakano, S., Eso, Y., Okada, H., et al. (2020) Recent Advances in Immunotherapy for Hepatocellular Carcinoma. Cancers, 12, 775. https://doi.org/10.3390/cancers12040775
- 2. European Association for the Study of the Liver (2018) EASL Clinical Practice Guidelines: Management of Hepatocellular Carcinoma. Journal of Hepatology, 69, 182-236. https://doi.org/10.1016/j.jhep.2018.03.019
- 3. Zheng, Z., Liang, W., Wang, D., et al. (2015) Adjuvant Chemotherapy for Patients with Primary Hepatocellular Carcinoma: A Meta-Analysis. International Journal of Cancer, 136, E751-E759. https://doi.org/10.1002/ijc.29203
- 4. Hargadon, K.M., Johnson, C.E. and Williams, C.J. (2018) Immune Checkpoint Blockade Therapy for Cancer: An Overview of FDA-Approved Immune Checkpoint Inhibitors. International Immunopharmacology, 62, 29-39. https://doi.org/10.1016/j.intimp.2018.06.001
- 5. Van Allen, E.M., Miao, D., Schilling, B., et al. (2015) Genomic Correlates of Response to CTLA-4 Blockade in Metastatic Melanoma. Science (New York, N.Y.), 350, 207-211. https://doi.org/10.1126/science.aad0095
- 6. Rizvi, N.A., Hellmann, M.D., Snyder, A., et al. (2015) Cancer Immunology. Mutational Landscape Determines Sensitivity to PD-1 Blockade in Non-Small Cell Lung Cancer. Science (New York, N.Y.), 348, 124-128.
- 7. Hugo, W., Zaretsky, J.M., Sun, L., et al. (2017) Genomic and Transcriptomic Features of Response to Anti-PD-1 Therapy in Metastatic Melanoma. Cell, 168, 542. https://doi.org/10.1016/j.cell.2017.01.010
- 8. Matsushita, H., Vesely, M.D., Koboldt, D.C., et al. (2012) Cancer Exome Analysis Reveals a T-Cell-Dependent Mechanism of Cancer Immunoediting. Nature, 482, 400-404. https://doi.org/10.1038/nature10755
- 9. Riaz, N., Morris, L., Havel, J.J., et al. (2016) The Role of Neoantigens in Response to Immune Checkpoint Blockade. International Immunology, 28, 411-419. https://doi.org/10.1093/intimm/dxw019
- 10. Coulie, P.G., Van den Eynde, B.J., van der Bruggen, P. and Boon, T. (2014) Tumour Antigens Recognized by T Lymphocytes: At the Core of Cancer Immunotherapy. Nature Reviews Cancer, 14, 135-146. https://doi.org/10.1038/nrc3670
- 11. Carreno, B.M., Magrini, V., Becker-Hapak, M., et al. (2015) Cancer Immunotherapy. A Dendritic Cell Vaccine Increases the Breadth and Diversity of Melanoma Neoantigen-Specific T Cells. Science (New York, N.Y.), 348, 803-808. https://doi.org/10.1126/science.aaa3828
- 12. Snyder, A. and Chan, T.A. (2015) Immunogenic Peptide Discovery in Cancer Genomes. Current Opinion in Genetics & Development, 30, 7-16. https://doi.org/10.1016/j.gde.2014.12.003
- 13. Ashburner, M., Ball, C.A., Blake, J.A., et al. (2000) Gene Ontology: Tool for the Unification of Biology. The Gene Ontology Consortium. Nature Genetics, 25, 25-29. https://doi.org/10.1038/75556
- 14. Kanehisa, M., Sato, Y., Kawashima, M., et al. (2016) KEGG as a Reference Resource for Gene and Protein Annotation. Nucleic Acids Research, 44, D457-D462. https://doi.org/10.1093/nar/gkv1070
- 15. Yu, G., Wang, L., Han, Y. and He, Q. (2012) clusterProfiler: An R Package for Comparing Biological Themes among Gene Clusters. OMICS, 16, 284-287. https://doi.org/10.1089/omi.2011.0118
- 16. Szklarczyk, D., Franceschini, A., Wyder, S., et al. (2015) STRING v10: Protein-Protein Interaction Networks, Integrated over the Tree of Life. Nucleic Acids Research, 43, D447-D452. https://doi.org/10.1093/nar/gku1003
- 17. Shannon, P., Markiel, A., Ozier, O., et al. (2003) Cytoscape: A Software Environment for Integrated Models of Biomolecular Interaction Networks. Genome Research, 13, 2498-2504. https://doi.org/10.1101/gr.1239303
- 18. Chin, C.-H., Chen, S.-H., Wu, H.-H., et al. (2014) cytoHubba: Identifying Hub Objects and Sub-Networks from Complex Interactome. BMC Systems Biology, 8, Article No. S11. https://doi.org/10.1186/1752-0509-8-S4-S11
- 19. Chen, B., Khodadoust, M.S., Liu, C.L., et al. (2018) Profiling Tumor Infiltrating Immune Cells with CIBERSORT. In: von Stechow, L., Ed., Cancer Systems Biology. Methods in Molecular Biology, Vol. 1711, Humana Press, New York, 243-259. https://doi.org/10.1007/978-1-4939-7493-1_12
- 20. Chan, T.A., Yarchoan, M., Jaffee, E., et al. (2019) Development of Tumor Mutation Burden as an Immunotherapy Biomarker: Utility for the Oncology Clinic. Annals of Oncology, 30, 44-56. https://doi.org/10.1093/annonc/mdy495
- 21. Lawrence, M.S., Stojanov, P., Polak, P., et al. (2013) Mutational Heterogeneity in Cancer and the Search for New Cancer-Associated Genes. Nature, 499, 214-218. https://doi.org/10.1038/nature12213
- 22. Snyder, A., Makarov, V., Merghoub, T., et al. (2014) Genetic Basis for Clinical Response to CTLA-4 Blockade in Melanoma. The New England Journal of Medicine, 371, 2189-2199. https://doi.org/10.1056/NEJMoa1406498
- 23. Chalmers, Z.R., Connelly, C.F., Fabrizio, D., et al. (2017) Analysis of 100,000 Human Cancer Genomes Reveals the Landscape of Tumor Mutational Burden. Genome Medicine, 9, Article No. 34. https://doi.org/10.1186/s13073-017-0424-2
- 24. Innocenti, F., Ou, F., Qu, X., et al. (2019) Mutational Analysis of Patients with Colorectal Cancer in CALGB/SWOG 80405 Identifies New Roles of Microsatellite Instability and Tumor Mutational Burden for Patient Outcome. Journal of Clinical Oncology, 37, 1217-1227. https://doi.org/10.1200/JCO.18.01798
- 25. Wang, X. and Li, M. (2019) Correlate Tumor Mutation Burden with Immune Signatures in Human Cancers. BMC Immunology, 20, Article No. 4. https://doi.org/10.1186/s12865-018-0285-5
- 26. Wu, T. and Dai, Y. (2017) Tumor Microenvironment and Therapeutic Response. Cancer Letters, 387, 61-68. https://doi.org/10.1016/j.canlet.2016.01.043
- 27. Hanahan, D. and Coussens, L.M. (2012) Accessories to the Crime: Functions of Cells Recruited to the Tumor Microenvironment. Cancer Cell, 21, 309-322. https://doi.org/10.1016/j.ccr.2012.02.022
- 28. Quail, D.F. and Joyce, J.A. (2013) Microenvironmental Regulation of Tumor Progression and Metastasis. Nature Medicine, 19, 1423-1437. https://doi.org/10.1038/nm.3394
- 29. De Plaen, E., Traversari, C., Gaforio, J.J., et al. (1994) Structure, Chromosomal Localization, and Expression of 12 Genes of the MAGE Family. Immunogenetics, 40, 360-369. https://doi.org/10.1007/BF01246677
- 30. Saito, T., Wada, H., Yamasaki, M., et al. (2014) High Expression of MAGE-A4 and MHC Class I Antigens in Tumor Cells and Induction of MAGE-A4 Immune Responses Are Prognostic Markers of CHP-MAGE-A4 Cancer Vaccine. Vaccine, 32, 5901-5907. https://doi.org/10.1016/j.vaccine.2014.09.002
- 31. Rapoport, A.P., Aqui, N.A., Stadtmauer, E.A., et al. (2014) Combination Immunotherapy after ASCT for Multiple myeloma Using MAGE-A3/Poly-ICLC Immunizations Followed by Adoptive Transfer of Vaccine-Primed and Costimulated Autologous T Cells. Clinical Cancer Research, 20, 1355-1365. https://doi.org/10.1158/1078-0432.CCR-13-2817
- 32. Mongkhoune, S., Xie, Y.A., Wang, Y.Q., et al. (2013) A Constructed HLA-A2-Restricted pMAGE-A1(278-286) Tetramer Detects Specific Cytotoxic T Lymphocytes in Tumour Tissues in Situ. The Journal of International Medical Research, 41, 1811-1824. https://doi.org/10.1177/0300060513496187
- 33. Makise, N., Morikawa, T., Nakagawa, T., et al. (2016) MAGE-A Expression, Immune Microenvironment, and Prognosis in Upper Urinary Tract Carcinoma. Human Pathology, 50, 62-69. https://doi.org/10.1016/j.humpath.2015.11.007
- 34. Cui, Z., Yu, X., Guo, L., et al. (2013) Combined Analysis of Serum Alpha-Fetoprotein and MAGE-A3-Specific Cytotoxic T Lymphocytes in Peripheral Blood for Diagnosis of Hepatocellular Carcinoma. Disease Markers, 35, Article ID: 907394. https://doi.org/10.1155/2013/907394
- 35. Babiak, A., Steinhauser, M., Götz, M., et al. (2014) Frequent T Cell Responses against Immunogenic Targets in Lung Cancer Patients for Targeted Immunotherapy. Oncology Reports, 31, 384-390. https://doi.org/10.3892/or.2013.2804
- 36. Balkwill, F. (2003) Chemokine Biology in Cancer. Seminars in Immunology, 15, 49-55. https://doi.org/10.1016/S1044-5323(02)00127-6
- 37. Borst, J., Ahrends, T., Bąbała, N., et al. (2018) CD4+ T Cell Help in Cancer Immunology and Immunotherapy. Nature Reviews Immunology, 18, 635-647. https://doi.org/10.1038/s41577-018-0044-0
- 38. Melssen, M. and Slingluff Jr., C.L. (2017) Vaccines Targeting Helper T Cells for Cancer Immunotherapy. Current Opinion in Immunology, 47, 85-92. https://doi.org/10.1016/j.coi.2017.07.004
- 39. Kennedy, R. and Celis, E. (2008) Multiple Roles for CD4+ T Cells in Anti-Tumor Immune Responses. Immunological Reviews, 222, 129-144. https://doi.org/10.1111/j.1600-065X.2008.00616.x
- 40. Bevan, M.J. (2004) Helping the CD8+ T-Cell Response. Nature Reviews Immunology, 4, 595-602. https://doi.org/10.1038/nri1413
- 41. Castellino, F. and Germain, R.N. (2006) Cooperation between CD4+ and CD8+ T Cells: When, Where, and How. Annual Review of Immunology, 24, 519-540. https://doi.org/10.1146/annurev.immunol.23.021704.115825
- 42. Ahrends, T., Spanjaard, A., Pilzecker, B., et al. (2017) CD4 T Cell Help Confers a Cytotoxic T Cell Effector Program Including Coinhibitory Receptor Downregulation and Increased Tissue Invasiveness. Immunity, 47, 848-861.E5. https://doi.org/10.1016/j.immuni.2017.10.009