Advances in Clinical Medicine
Vol.
11
No.
03
(
2021
), Article ID:
41215
,
11
pages
10.12677/ACM.2021.113180
基于生物信息学方法筛选影响胶质母细胞瘤的诊断及治疗的关键基因
肖珂,代兴亮,茆翔,高鹏,周律,程宏伟*
安徽医科大学第一附属医院神经外科,安徽 合肥

收稿日期:2021年2月17日;录用日期:2021年3月5日;发布日期:2021年3月25日

摘要
目的:通过生物信息学方法对胶质母细胞瘤公共数据库进行信息挖掘以筛选胶质母细胞瘤的关键差异表达基因,预测胶质母细胞瘤的可能潜在治疗靶点基因。方法:从基因表达数据库(GEO)中下载脑胶质母细胞瘤数据,通过在线分析软件GEO2R筛选正常组织与胶质母细胞瘤组织差异表达基因。通过Venn图取三个差异基因集交集。共筛选出632个共有差异基因。再通过STRING网站里的在线分析软件绘制PPI (protein-protein interaction network,蛋白互作网络),并导入Cytoscape软件利用MCODE插件筛选关键基因。通过R软件的Goplot包对差异表达基因进行GO和KEGG功能富集分析。再通过UCSC网站的癌症基因浏览器获得各关键基因在脑胶质母细胞瘤中的表达。通过GEPIA2网站绘制各关键基因的生存曲线。通过Oncomine网站获得关键预后基因KDELR2、CDKN3及RELN基因在不同级别胶质瘤中的表达。结果:从GEO数据库中下载GSE104291、GSE15824、GSE66354的数据集,通过GEO2R分析出差异表达基因,再通过Venn图共筛选出632个共有差异表达基因。通过STRING网站构建蛋白互作网络,并导入Cytoscape软件筛选出关键基因。获得差异表达基因的GO及KEGG功能富集分析图。通过GEPIA2网站获得关键基因的总生存率及无病生存率,结果发现KDELR2、CDKN3及RELN基因的表达差异与总生存率(OS)和无病生存率(RFS)相关性具有统计学意义(KDELR2、CDKN3和RELN这三个基因的总生存率生存曲线Logrank P值分别为<0.001,<0.001以及1.7e−10;KDELR2、CDKN3和RELN这三个基因的无病生存率生存曲线Logrank P值分别为<0.001,<0.001以及0.00032)。KDELR2及CDKN3在正常组织中低表达,在胶质瘤组织表达与级别成正相关;RELN在在正常组织中高表达,在胶质瘤组织表达与级别成负相关。结论:通过本研究发现胶质瘤OS和RFS与KDELR2及CDKN3基因呈负相关,与RELN基因呈正相关。结合这些基因的表达差异,由此推测KDELR2基因及CDKN3基因可能是脑胶质母细胞瘤的发生及进展的癌基因,而RELN基因可能是一个抑癌基因。
关键词
胶质母细胞瘤,生物信息分析,KDELR2,CDKN3,RELN
Screening of Key Genes Affecting the Diagnosis and Treatment of Glioblastoma Based on Bioinformatics
Ke Xiao, Xingliang Dai, Xiang Mao, Peng Gao, Lv Zhou, Hongwei Cheng*
Department of Neurosurgery, The First Affiliated Hospital of Anhui Medical University, Hefei Anhui

Received: Feb. 17th, 2021; accepted: Mar. 5th, 2021; published: Mar. 25th, 2021

ABSTRACT
Objective: To mine the information of glioblastoma public database by bioinformatics method to screen the key differentially expressed genes of glioblastoma and to predict the potential therapeutic target genes of glioblastoma. Methods: The glioblastoma data were downloaded from the Gene Expression Omnibus database (GEO), and the differentially expressed genes between normal and glioblastoma tissues were screened by online analysis software GEO2R. The intersection of three differential gene sets was obtained by Venn map. A total of 632 common differential genes were screened. Then draw PPI (protein-protein interaction network) through the online analysis software in STRING website, and then import the Cytoscape software to screen key genes using MCODE plug-ins. The functional enrichment of differentially expressed genes was analyzed by GO and KEGG by Goplot package of R software. Then the expression of key genes in glioblastoma was obtained by cancer gene browser on UCSC website. The survival curve of key genes was drawn by GEPIA2 website. The expressions of key prognostic genes KDELR2, CDKN3 and RELN in different grades of gliomas were obtained by Oncomine website. Results: The data sets of GSE104291, GSE15824 and GSE66354 were downloaded from GEO database, and the differentially expressed genes were analyzed by GEO2R. A total of 632 common differentially expressed genes were screened by Venn map. The PPI network was constructed by STRING website, and the key genes were screened by Cytoscape software. GO and KEGG functional enrichment analysis maps of differentially expressed genes were obtained. The overall survival rate and disease-free survival rate of key genes were obtained by GEPIA2 website. The results showed that the expression differences of KDELR2, CDKN3 and RELN genes were significantly correlated with overall survival rate (OS) and disease-free survival rate (RFS) (the Logrank P values of the overall survival curves of KDELR2, CDKN3 and RELN were <0.001, <0.001 and 1.7e−10, respectively; the Logrank P values of the disease-free survival curves of KDELR2, CDKN3 and RELN were <0.001, <0.001 and 0.00031, respectively). The expression of KDELR2 and CDKN3 was low in normal tissues, but positively correlated with grade in glioma tissues, while RELN was highly expressed in normal tissues and negatively correlated with grade in glioma tissues. Conclusion: It is found that OS and RFS in gliomas are negatively correlated with KDELR2 and CDKN3 genes, and positively correlated with RELN genes. Combined with the differences in the expression of these genes, it is speculated that KDELR2 gene and CDKN3 gene may be oncogenes in the occurrence and progression of glioblastoma, while RELN gene may be a tumor suppressor gene.
Keywords:Glioblastoma, Bioinformatics, KDELR2, CDKN3, RELN

Copyright © 2021 by author(s) and Hans Publishers Inc.
This work is licensed under the Creative Commons Attribution International License (CC BY 4.0).
http://creativecommons.org/licenses/by/4.0/


1. 引言
胶质母细胞瘤是人类脑恶性肿瘤当中最常见的肿瘤,恶性程度最高,5年总生存率极低 [1]。目前胶质瘤的发病机制仍然未完全阐明,可能与多种基因的突变共同作用有关。胶质母细胞瘤作为胶质瘤级别最高的肿瘤,其中位生存期是14.6个月,五年生存率仅9.8% [2]。胶质母细胞瘤现在最主要的经典治疗方案仍然是手术尽量全切,接着伴随着同步放化疗 [3]。但是由于胶质母细胞瘤的高度侵袭浸润性,近乎100%的复发率仍然是困扰我们的难题。目前靶向治疗对于恶性肿瘤来说是一个划时代的治疗方案,因其高度精准、疗效显著而受到广大临床工作者及患者的青睐。胶质母细胞瘤的靶向治疗依然是现在研究的热点。问题的关键在于靶点靶基因的选择,既往筛选工作耗时耗力,现如今通过生物信息学方法,可以较迅速地获得胶质母细胞瘤的关键差异表达基因,并对这些可能成为靶点基因的基因进行下一步研究。对于胶质母细胞瘤,寻找到敏感度和特异度都比较高的分子标记物可以帮助我们提高诊断的几率,进一步提高我们的治疗率。本文通过生物信息学方法筛选差异基因,可能提高我们对于胶质瘤的诊断率,作为我们治疗胶质母细胞瘤的靶点,进一步影响预后。对于肿瘤的发生机制也可以深化研究。
2. 资料与方法
2.1. 资料来源
通过NCBI的GEO数据库中下载胶质母细胞瘤的基因表达谱的GSE104291、GSE15824、GSE66354的数据集。然后利用在线分析软件GEO2R筛选正常脑组织与胶质母细胞瘤组织差异表达基因。
2.2. 方法
2.2.1. 数据的处理及差异表达基因的筛选
GEO数据库是一个包含基因表达数据、芯片、微阵列的公共公开的数据库 [4]。我们主要从基因表达数据库(GEO)中下载脑胶质母细胞瘤基因表达数据,通过在线分析软件GEO2R筛选正常组织与胶质瘤组织差异表达基因。同时要求筛选到的差异基因log2FC的绝对值大于2。校正后的P值小于0.01。再将符合要求的基因导入bioinformatics.psb.ugent.be网站通过Venn图工具取三个差异基因集交集,以进一步聚焦其中显著的差异表达基因。
2.2.2. 构建PPI网络
PPI网络(protein-protein interaction network,蛋白互作网络)的构建需要借用STRING数据库(https://string-db.org)的搜索工具 [5]。蛋白互作网络可以揭露各个基因或蛋白之间的关系,进一步筛选出其中的关键差异表达基因。将筛选到的差异基因通过STRING网站里的在线分析软件绘制PPI (蛋白互作网络)。我们选择的是Multiple proteins,要求的组织是Homo sapiens。
2.2.3. 获得关键基因并对差异表达基因进行GO和KEGG富集分析
Cytoscape软件是一个使分子干预网络可视化的开源的生物信息软件 [6]。我们首先将获得的蛋白互作网络图导入到Cytoscape,通过Cytoscape软件中的MCODE插件,筛选其中的关键基因 [7]。R语言的Goplot包可以进行基因的功能富集分析。KEGG是一个数据库资源,用于从高通量实验技术产生的大规模分子数据集中了解基因的高级功能和生物学系统 [8]。GO是一种重要的生物信息学工具,用于注释基因和分析这些基因的生物学过程 [9]。我们将差异表达基因通过R语言的Goplot包获得差异表达基因的GO和KEGG通路富集分析,富集条件为:物种为人类(human);基因为官方名称。这样就可以获得这些差异表达基因富集最多的功能,以进一步加深对这些差异表达基因的理解。
2.2.4. 获得各关键基因在脑胶质瘤组织中的表达
将关键基因导入到UCSC (http://genome-cancer.ucsc.edu)的癌症基因浏览器获得关键基因在胶质母细胞瘤组织中的基因表达量 [10]。
2.2.5. 关键基因的生存曲线的获得
GEPIA2是北京大学张泽民老师实验室开发的一个网站,能够对TCGA和GTEx项目共9736个肿瘤样本、8587个正常样本的RNA-seq表达数据进行分析 [11]。通过GEPIA2网站(http://gepia2.cancer-pku.cn/)绘制各关键基因在胶质瘤中的生存曲线,主要包括总生存率(OS, Overall Survival)和无病生存率(RFS, Disease Free Survival) [12] [13],以筛选出其中与预后相关的关键差异表达基因。
2.2.6. KDELR2、CDKN3、RELN基因在正常组织和胶质瘤组织中的表达
Oncomine网站(http://www.oncomine.com)集合了大量的各种肿瘤的基因表达及临床数据。通过Oncomine网站来分析KDELR2、CDKN3、RELN基因在正常组织和不同级别的胶质瘤组织中的表达 [14] [15] [16]。
3. 结果
3.1. 数据的处理及差异表达基因的筛选
我们从GEO数据库中的GSE104291数据集中筛选出2028个差异表达基因,从GSE15824中筛选出3237个差异表达基因,从GSE66354数据集中筛选出2164个差异表达基因,再将这三个数据集中筛选出的差异基因导入Venn图软件,共获得三个数据集共有差异表达基因632个。见图1。

Figure 1. Venn diagram of three gene sets
图1. 三个基因集的韦恩图
3.2. 构建PPI网络
我们将获得的这632个差异基因导入到STRING数据库获得蛋白互作网络(PPI)见图2。从图中可以发现各基因表达产物蛋白质之间的关系。
3.3. 获得关键基因并对差异表达基因进行GO和KEGG富集分析
将获得的PPI网络导入Cytoscape软件。通过Cytoscape软件中的MCODE插件筛选到关键基因11个,他们分别为CDKN3、GRM5、OPALIN、CAMK2B、SHANK2、RELN、CDK2、KDELR2、NTNG2、FUT9、HS3ST4。利用R语言的Goplot包获得的差异表达基因GO和KEGG富集分析见图3。GO主要包括三个方面进行基因注释,即:生物过程(BP)、细胞组分(CC)、分子功能(MF)。由于篇幅有限,我们分别将生物过程、细胞组分、分子功能取富集最多的前10个项目可视化在图3(a)。取KEGG通路最显著的前20个项目可视化在图3(b)。从图中我们可以发现在BP中这些差异表达基因富集最多的在突触传递的化学调节,在CC中主要集中在突触膜中,在MF中则主要在离子通道中集中。而最集中的KEGG通路是在神经活动配体–受体相互作用通路和γ-氨基丁酸能突触通路。

Figure 2. PPI network of 632 differentially expressed genes
图2. 632个差异表达基因蛋白互作网络(PPI)
3.4. 获得各关键基因在脑胶质瘤组织中的表达
将这11个关键基因输入UCSC网站癌症基因浏览器,以获得各关键基因在胶质母细胞瘤组织中的表达量。见图4。
 (a)
(a)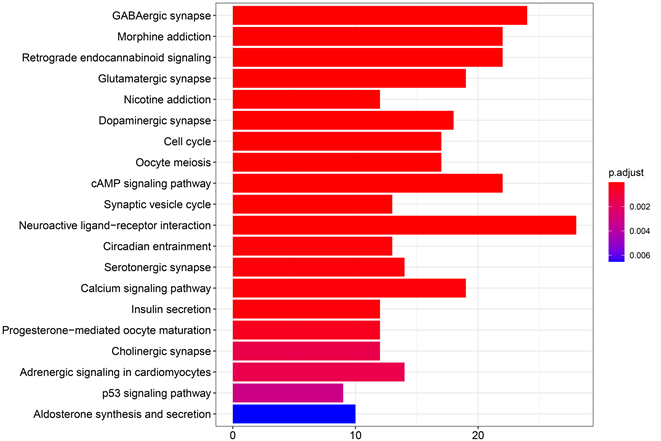 (b)
(b)
Figure 3. Functional enrichment analysis of GO and KEGG of key differentially expressed genes
图3. 关键差异表达基因的GO和KEGG功能富集分析
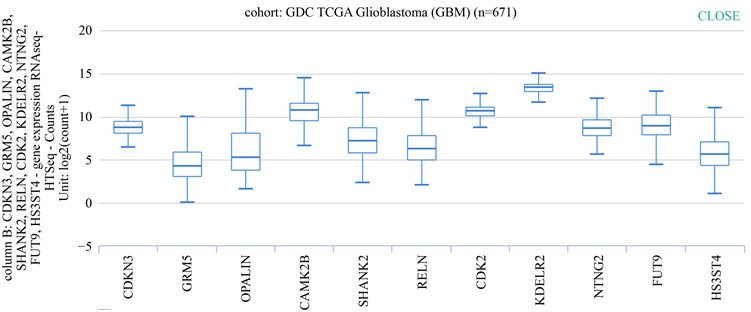
Figure 4. Expression of key differentially expressed genes in glioblastoma
图4. 关键差异表达基因在胶质母细胞瘤的表达量
3.5. 关键基因的生存曲线的获得
将这11个关键基因通过GEPIA2网站绘制各关键基因的在胶质瘤中的生存曲线,最终发现KDELR2、 CDKN3和RELN这3个基因有统计学意义(Logrank P-Value < 0.05),与胶质瘤的预后相关。胶质瘤OS和RFS与KDELR2及CDKN3基因呈负相关,与RELN基因呈正相关。见图5。从图中我们可以发现


Figure 5. Survival analysis of key differentially expressed genes
图5. 关键差异表达基因的生存分析
KDELR2、CDKN3和RELN这三个基因的总生存率生存曲线Logrank P值分别为<0.001,<0.001以及1.7e−10;KDELR2、CDKN3和RELN这三个基因的无病生存率生存曲线Logrank P值分别为<0.001,<0.001以及0.00031。
3.6. KDELR2、CDKN3、RELN基因在正常组织和胶质瘤组织中的表达
我们将预后相关基因KDELR2、CDKN3、RELN基因导入Oncomine网站进一步探索他们在胶质瘤及正常脑组织中的表达,最终发现KDELR2及CDKN3在正常组织中低表达,在胶质瘤组织表达与级别成正相关;RELN在在正常组织中高表达,在胶质瘤组织表达与级别成负相关。所用到的数据库为Sun Brain数据库。见图6。

Figure 6. Expression of KDELR2, CDKN3 and RELN genes in normal and glioma tissues
图6. KDELR2、CDKN3、RELN基因在正常组织和胶质瘤组织中的表达
4. 讨论
胶质瘤仍然是困扰人类的一种极度恶性肿瘤之一。由于胶质母细胞瘤的高度侵袭性,胶质母细胞瘤的复发率近乎100% [17]。对于IV级胶质瘤(胶质母细胞瘤)患者,平均生存期小于15个月 [18]。一些患者对化疗和放疗并不敏感,很多会很快复发 [19]。目前对于胶质母细胞瘤仍然没有一种理想的特效化疗药,绝大多数胶质母细胞瘤瘤对化疗药不敏感。作为胶质母细胞瘤最经典的化疗药,替莫唑胺的耐药使得胶质母细胞瘤的治疗大打折扣 [20]。胶质母细胞瘤不像肝癌,有敏感度和特异度都比较高的分子标志物——AFP(甲胎蛋白) [21]。所以相对于肝癌,胶质母细胞瘤瘤的发现仍然主要依靠影像学,但头部影像学筛查并不是很普及。所以研究出一种灵敏度和特异度都比较高的分子标记物显得至关重要。
通过本研究可以发现KDELR2及CDKN3在正常脑组织中低表达,在胶质瘤级别越高组织中越高表达。我们获得的11个关键基因仅KDELR2、CDKN3和RELN与患者的预后有关。下一步我们可以进行一些关于这些基因的实验研究,比如:集落形成实验、划痕实验、流式细胞仪分析、实时荧光定量PCR、免疫共沉淀试验、染色质免疫沉淀试验等,以观察这些基因在胶质母细胞瘤细胞生长、上皮–间充质转化、胶质瘤干细胞的迁移、侵袭和生成等方面的作用。
CDKN3是CDK家族的重要组成,CDKN3编码的蛋白属于蛋白磷酸酶家族抑制因子。它的功能是在Thr160处使CDK2激酶去磷酸化,从而阻止CDK2的激活 [22] [23]。一些研究发现,CDKN3在多种肿瘤当中扮演着重要作用,比如食管癌、宫颈癌、鼻咽癌等 [24] [25] [26]。在胶质母细胞瘤细胞中,CDKN3可通过磷酸酶依赖性抑制CDC2抑制细胞增殖和迁移 [27]。本研究发现在胶质瘤中CDKN3扮演着重要角色,级别越高的胶质瘤,其表达量越高。KDELRs的主要功能是作为载体来取回多个伴随蛋白如GRP78从高尔基体到内质网。KDELRs和GRP78在内质网应激时均是上调的。GRP78是肿瘤发生的一个关键调控因子。KDELR2过度表达降低了GRP78在细胞表面的表达,但对GRP78的总体表达影响不大 [28]。对胶质瘤多基因表达谱的一个分析显示:KDELR2在胶质母细胞瘤和低级别胶质瘤中均是核心基因,KDELR2的表达可以将病人分为不同生存风险,KDELR2的高表达往往预示病人的生存期比较短,而KDELR2的低表达则预示着病人的生存期比较长 [29]。KDELR2基因的研究相对来说较少,为我们研究胶质母细胞瘤提供了一个新方向。RELN属于富含半胱氨酸的分泌蛋白家族。关于他的研究很少,但一些研究已经发现它与一些肿瘤有关,如:结肠癌 [30]。
总之,本研究发现KDELR2及CDKN3在胶质母细胞瘤的发生发展中扮演着重要的作用,其靶基因涉及胶质母细胞瘤的众多通路,为胶质母细胞瘤的诊断及靶向治疗等提供了方向。而RELN基因可能是一个抑癌基因。
5. 结论
本研究利用生物信息学方法,通过各种工具、网站,筛选出了关键预后相关基因:KDELR2,CDKN3,RELN。KDELR2基因及CDKN3基因可能是脑胶质母细胞瘤的发生及进展的癌基因,而RELN基因可能是一个抑癌基因。
文章引用
肖 珂,代兴亮,茆 翔,高 鹏,周 律,程宏伟. 基于生物信息学方法筛选影响胶质母细胞瘤的诊断及治疗的关键基因
Screening of Key Genes Affecting the Diagnosis and Treatment of Glioblastoma Based on Bioinformatics[J]. 临床医学进展, 2021, 11(03): 1250-1260. https://doi.org/10.12677/ACM.2021.113180
参考文献
- 1. Siegel, R.L., Miller, K.D. and Jemal, A. (2020) Cancer Statistics, 2020. CA: A Cancer Journal for Clinicians, 70, 7-30. https://doi.org/10.3322/caac.21590
- 2. Stupp, R., Hegi, M.E., Mason, W.P., van den Bent, M.J., Taphoorn, M.J., Janzer, R.C., et al. (2009) Effects of Radiotherapy with Concomitant and Adjuvant Temozolomide versus Radiotherapy Alone on Survival in Glioblastoma in a Randomised Phase III Study: 5-Year Analysis of the EORTC-NCIC Trial. The Lancet Oncology, 10, 459-466. https://doi.org/10.1016/S1470-2045(09)70025-7
- 3. Stupp, R., Mason, W.P., van den Bent, M.J., Weller, M., Fisher, B., Taphoorn, M.J., et al. (2005) Radiotherapy plus Concomitant and Adjuvant Temozolomide for Glioblastoma. The New England Journal of Medicine, 352, 987-996. https://doi.org/10.1056/NEJMoa043330
- 4. Edgar, R., Domrachev, M. and Lash, A.E. (2002) Gene Expression Omnibus: NCBI Gene Expression and Hybridization Array Data Repository. Nucleic Acids Research, 30, 207-210. https://doi.org/10.1093/nar/30.1.207
- 5. Franceschini, A., Szklarczyk, D., Frankild, S., Kuhn, M., Simonovic, M., Roth, A., et al. (2013) STRING v9.1: Protein-Protein Interaction Networks, with Increased Coverage and Integration. Nucleic Acids Research, 41, D808-D815. https://doi.org/10.1093/nar/gks1094
- 6. Smoot, M.E., Ono, K., Ruscheinski, J., Wang, P.L. and Ideker, T. (2011) Cytoscape 2.8: New Features for Data Integration and Network Visualization. Bioinformatics, 27, 431-432. https://doi.org/10.1093/bioinformatics/btq675
- 7. Bandettini, W.P., Kellman, P., Mancini, C., Booker, O.J., Vasu, S., Leung, S.W., et al. (2012) MultiContrast Delayed Enhancement (MCODE) Improves Detection of Subendocardial Myocardial Infarction by Late Gadolinium Enhancement Cardiovascular Magnetic Resonance: A Clinical Validation Study. Journal of Cardiovascular Magnetic Resonance, 14, 83. https://doi.org/10.1186/1532-429X-14-83
- 8. Kanehisa, M. (2002) The KEGG Database. Novartis Foundation Symposia, 247, 91-101, 101-103, 119-128, 244-252. https://doi.org/10.1002/0470857897.ch8
- 9. Ashburner, M., Ball, C.A., Blake, J.A., Botstein, D., Butler, H., Cherry, J.M., et al. (2000) Gene Ontology: Tool for the Unification of Biology. The Gene Ontology Consortium. Nature Genetics, 25, 25-29. https://doi.org/10.1038/75556
- 10. Haeussler, M., Zweig, A.S., Tyner, C., Speir, M.L., Rosenbloom, K.R., Raney, B.J., et al. (2019) The UCSC Genome Browser Database: 2019 Update. Nucleic Acids Research, 47, D853-D858. https://doi.org/10.1093/nar/gky1095
- 11. Tang, Z., Kang, B., Li, C., Chen, T. and Zhang, Z. (2019) GEPIA2: An Enhanced Web Server for Large-Scale Expression Profiling and Interactive Analysis. Nucleic Acids Research, 47, W556-W560. https://doi.org/10.1093/nar/gkz430
- 12. Gao, J., Aksoy, B.A., Dogrusoz, U., Dresdner, G., Gross, B., Sumer, S.O., et al. (2013) Integrative Analysis of Complex Cancer Genomics and Clinical Profiles Using the cBioPortal. Science Signaling, 6, l1. https://doi.org/10.1126/scisignal.2004088
- 13. Cerami, E., Gao, J., Dogrusoz, U., Gross, B.E., Sumer, S.O., Aksoy, B.A., et al. (2012) The cBio Cancer Genomics Portal: An Open Platform for Exploring Multidimensional Cancer Genomics Data. Cancer Discovery, 2, 401-404. https://doi.org/10.1158/2159-8290.CD-12-0095
- 14. Chen, X., Cheung, S.T., So, S., Fan, S.T., Barry, C., Higgins, J., et al. (2002) Gene Expression Patterns in Human Liver Cancers. Molecular Biology of the Cell, 13, 1929-1939. https://doi.org/10.1091/mbc.02-02-0023
- 15. Roessler, S., Jia, H.L., Budhu, A., Forgues, M., Ye, Q.H., Lee, J.S., et al. (2010) A Unique Metastasis Gene Signature Enables Prediction of Tumor Relapse in Early-Stage Hepatocellular Carcinoma Patients. Cancer Research, 70, 10202-10212. https://doi.org/10.1158/0008-5472.CAN-10-2607
- 16. Wurmbach, E., Chen, Y.B., Khitrov, G., Zhang, W., Roayaie, S., Schwartz, M., et al. (2007) Genome-Wide Molecular Profiles of HCV-Induced Dysplasia and Hepatocellular Carcinoma. Hepatology, 45, 938-947. https://doi.org/10.1002/hep.21622
- 17. Louis, D.N., Perry, A., Reifenberger, G., von Deimling, A., Figarella-Branger, D., Cavenee, W.K., et al. (2016) The 2016 World Health Organization Classification of Tumors of the Central Nervous System: A Summary. Acta Neuropathologica, 131, 803-820. https://doi.org/10.1007/s00401-016-1545-1
- 18. Rasmussen, B.K., Hansen, S., Laursen, R.J., Kosteljanetz, M., Schultz, H., Norgard, B.M., et al. (2017) Epidemiology of Glioma: Clinical Characteristics, Symptoms, and Predictors of Glioma Patients Grade I-IV in the Danish Neuro-Oncology Registry. Journal of Neuro-Oncology, 135, 571-579. https://doi.org/10.1007/s11060-017-2607-5
- 19. Lai, S.W., Huang, B.R., Liu, Y.S., Lin, H.Y., Chen, C.C., Tsai, C.F., et al. (2018) Differential Characterization of Temozolomide-Resistant Human Glioma Cells. International Journal of Molecular Sciences, 19, 127. https://doi.org/10.3390/ijms19010127
- 20. Lee, S.Y. (2016) Temozolomide Resistance in Glioblastoma Multiforme. Genes and Diseases, 3, 198-210. https://doi.org/10.1016/j.gendis.2016.04.007
- 21. Sauzay, C., Petit, A., Bourgeois, A.M., Barbare, J.C., Chauffert, B., Galmiche, A., et al. (2016) Alpha-Foetoprotein (AFP): A Multi-Purpose Marker in Hepatocellular Carcinoma. Clinica Chimica Acta, 463, 39-44. https://doi.org/10.1016/j.cca.2016.10.006
- 22. Gyuris, J., Golemis, E., Chertkov, H. and Brent, R. (1993) Cdi1, a Human G1 and S Phase Protein Phosphatase That Associates with Cdk2. Cell, 75, 791-803. https://doi.org/10.1016/0092-8674(93)90498-F
- 23. Hannon, G.J., Casso, D. and Beach, D. (1994) KAP: A Dual Specificity Phosphatase That Interacts with Cyclin-Dependent Kinases. Proceedings of the National Academy of Sciences of the United States of America, 91, 1731-1735. https://doi.org/10.1073/pnas.91.5.1731
- 24. Barron, E.V., Roman-Bassaure, E., Sanchez-Sandoval, A.L., Espinosa, A.M., Guardado-Estrada, M., Medina, I., et al. (2015) CDKN3 mRNA as a Biomarker for Survival and Therapeutic Target in Cervical Cancer. PLoS ONE, 10, e137397. https://doi.org/10.1371/journal.pone.0137397
- 25. Wang, J., Che, W., Wang, W., Su, G., Zhen, T. and Jiang, Z. (2019) CDKN3 Promotes Tumor Progression and Confers Cisplatin Resistance via RAD51 in Esophageal Cancer. Cancer Management and Research, 11, 3253-3264. https://doi.org/10.2147/CMAR.S193793
- 26. Wang, H., Chen, H., Zhou, H., Yu, W. and Lu, Z. (2017) Cyclin-Dependent Kinase Inhibitor 3 Promotes Cancer Cell Proliferation and Tumorigenesis in Nasopharyngeal Carcinoma by Targeting p27. Oncology Research, 25, 1431-1440. https://doi.org/10.3727/096504017X14835311718295
- 27. Nalepa, G., Barnholtz-Sloan, J., Enzor, R., Dey, D., He, Y., Gehlhausen, J.R., et al. (2013) The Tumor Suppressor CDKN3 Controls Mitosis. Journal of Cell Biology, 201, 997-1012. https://doi.org/10.1083/jcb.201205125
- 28. Tiwarekar, V., Fehrholz, M. and Schneider-Schaulies, J. (2019) KDELR2 Competes with Measles Virus Envelope Proteins for Cellular Chaperones Reducing Their Chaperone-Mediated Cell Surface Transport. Viruses, 11, 27. https://doi.org/10.3390/v11010027
- 29. Hsu, J.B., Chang, T., Lee, G.A., Lee, T.Y. and Chen, C.Y. (2019) Identification of Potential Biomarkers Related to Glioma Survival by Gene Expression Profile Analysis. BMC Medical Genomics, 11, 34. https://doi.org/10.1186/s12920-019-0479-6
- 30. Zheng, L.D., Tong, Q.S., Weng, M.X., He, J., Lv, Q., Pu, J.R., et al. (2009) Enhanced Expression of Resistin-Like Molecule Beta in Human Colon Cancer and Its Clinical Significance. Digestive Diseases and Sciences, 54, 274-281. https://doi.org/10.1007/s10620-008-0355-2
NOTES
*通讯作者。