Botanical Research
Vol.07 No.02(2018), Article ID:24132,6
pages
10.12677/BR.2018.72021
The Research Progress of Heat Shock Transcription Factors in Plants
Mingxuan Zhao, Xiaojun Hu*
Linyi University, Linyi Shandong

Received: Feb. 28th, 2018; accepted: Mar. 14th, 2018; published: Mar. 23rd, 2018

ABSTRACT
High temperature is often predicted to reduce crop yield and grain quality recently. Heat shock transcription factors (Hsfs) play a central regulatory role in alleviating the harm caused by high temperature on plant. Hsfs exist in all eukaryotes. According to the different characteristics of structure, Hsfs are divided into 3 classes, A, B and C. Class A and B genes are heat responsible, but Class C genes have no response to heat stress. Class As are reported to play a central regulation role in heat protection through regulating HSPs. But the biological roles of Class Bs are not well known, Class C, not known. Hsfs are reported to regulate not only in the process of heat tolerance but also drought, salt, oxidation, heavy metal and osmotic stress tolerance. A series of genes encoding heat shock proteins, molecular chaperone, active oxygen scavenging enzymes and other functional proteins can be transactivated by Hsfs. And the transactivation relies on the binding of the DNA binding domain and heat shock element (HSE) in the promoter regions of the down-stream genes. In this paper, the classification, biological function, down-stream genes and regulation mechanism of Hsfs are introduced, for the purpose of providing theoretical guidance to plant heat and other adverse stress research.
Keywords:High Temperature, Heat Shock Transcription Factors, Classification, Heat Shock Proteins, Regulation Mechanism
植物热激转录因子研究进展
赵明宣,胡晓君*
临沂大学,山东 临沂

收稿日期:2018年2月28日;录用日期:2018年3月14日;发布日期:2018年3月23日

摘 要
近年来高温天气带来的作物减产现象频现,热激转录因子家族Hsfs在缓解高温对植物的伤害中发挥重要的作用。Hsfs基因广泛存在于所有真核生物中,根据结构特点,Hsfs被分为A、B、C三大家族,目前对HsfA家族功能研究较多,对HsfB家族研究较少,对HsfC家族的研究未见报到。根据以往研究,Hsfs不仅参与植物抵御高温胁迫的调控过程,还参与植物抗旱、抗盐、抗氧化、抗重金属和抗高渗透胁迫过程。大部分Hsfs家族成员是通过调控一系列热激蛋白,分子伴侣,活性氧清除酶和其他功能蛋白基因发挥抗逆作用的。Hsfs通过与下游基因启动子区域的HSEs元件结合而调控下游基因的表达。本文综合了近几年的国内外研究报到,详细介绍了Hsfs家族的分类、功能、下游基因种类、调控机理等,以期为植物抗高温和其他逆境胁迫研究提供理论指导。
关键词 :高温,热激转录因子,分类,热激蛋白,调控机制

Copyright © 2018 by authors and Hans Publishers Inc.
This work is licensed under the Creative Commons Attribution International License (CC BY).
http://creativecommons.org/licenses/by/4.0/


1. 引言
随着全球气候变暖,高温胁迫带来的作物减产现象频现 [1] [2] 。特别是开花和灌浆期的高温胁迫,是导致谷类作物减产的主要原因之一 [1] [2] 。高温胁迫对植物的直接伤害是蛋白质分子变性、生物膜结构破损、自由基和活性氧化物累积、体内生理生化代谢紊乱 [3] 。植物体内存在着以Cbf/Dreb、bZIP、Myb/Myc、Wrky、Nac和Hsf等转录因子家族为核心的应对逆境胁迫的一系列信号感知传导基因网络调控系统,其中,热激转录因子家族Hsfs在缓解高温对植物的伤害中发挥重要作用 [4] 。
2. 热激转录因子的的种类
Hsfs基因存在于所有的真核生物中,酵母和动物体内只有少数Hsfs基因,而植物体内存在着大量的Hsfs基因 [3] 。到目前为止,至少分离鉴定了21个拟南芥Hsfs [5] 、16个番茄Hsfs [6] ,28个大麦Hsfs [7] ,25个水稻Hsfs [8] [9] ,34个大豆Hsfs [10] ,40个棉花Hsfs [11] 和56个小麦Hsfs [12] 。所有的Hsfs都具有一个高度保守的DNA结合结构域(DBD)和两个疏水氨基酸重复片段HR-A区和HR-B区 [3] [13] 。根据它们的蛋白质结构,Hsfs被分为A、B和C亚家族(图1)。HsfA亚家族在HR-A和HR-B之间有一段长的氨基酸链插入,在C末端含有一段富含芳香族–疏水–酸性氨基酸尾巴(AHA),HsfB亚家族在HR-A和HR-B之间没有插入序列,而HsfC亚家族成员在HR-A和HR-B之间的插入序列很短 [3] [13] 。近年来在单子叶植物中发现了一类特殊HsfC亚家族成员,被划分为HsfC2亚家族 [3] [13] ,HsfC2亚家族在HR-A和HR-B之间有很短的插入序列,在C末端有一个AHA相似的尾巴。
3. 热激转录因子的功能
Hsfs家族成员在植物抵御高温胁迫和其他逆境过程中发挥重要作用 [6] [14] [15] 。作为水稻,大豆,番茄和拟南芥中的主要Hsfs成员,超表达HsfA1显著提高它们的抗热性 [14] [15] 。超表达HsfA2基因显著
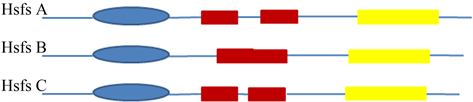 说明:图中蓝色椭圆部分为DNA结合域(DBD),红色框状部分为疏水氨基酸重复片段HR-A/B区域,黄色部分为芳香族-疏水-酸性氨基酸(AHA)尾巴。
说明:图中蓝色椭圆部分为DNA结合域(DBD),红色框状部分为疏水氨基酸重复片段HR-A/B区域,黄色部分为芳香族-疏水-酸性氨基酸(AHA)尾巴。
Figure 1. The classification of the Heat Shock Factors family
图1. 热激转录因子家族的分类
提高拟南芥的抗强光、抗盐、抗超氧化、抗高渗透胁迫的能力 [16] [17] [18] 。转基因拟南芥超表达水稻HsfA2e (p < 0.01)的抗盐性显著提高 [19] 而转基因拟南芥超表达番茄HsfA3 (p < 0.01)的盐敏性显著提高 [20] 。水稻或者小麦中超表达HsfA4a显著提高它们抗重金属元素镉的能力和抗氧化能力 [21] 。大部分HsfB亚家族成员的表达量在高温胁迫下显著升高,除此之外,HsfB亚家族成员被认为与生物胁迫的抗性调控有关,因为降低HsfB1和HsfB2b表达量引起植物真菌抗性基因表达量的增高 [22] [23] [24] 。同一转录因子在不同的植物中的功能也会有差异 [25] 。例如,HsfB1在番茄中被证明是一个协同激活因子,与HsfA家族成员一同参与热胁迫的调控 [26] ,而在拟南芥中,HsfB1被证明是HsfA2和HsfA7的抑制因子 [27] 。目前,HsfC亚家族基因的功能研究却很少被报道,特别是单子叶植物特异的HsfC2亚家族基因的功能尚未见报道。
4. 热激转录因子的下游基因
大部分Hsfs家族成员是通过调控一系列热激蛋白,分子伴侣,活性氧清除酶和其他功能蛋白基因发挥抗逆作用的,如HsfA1、A2、A3、A4、A6和A9等亚家族成员 [12] [25] 。转基因植物超表达这些Hsfs基因呈现出很强的抗热性,一系列的热激蛋白和高温保护有关的基因在转基因植株中的表达量增高,敲除这些Hsfs基因使植株对高温的敏感性提高 [6] [12] [23] [28] [29] 。转HsfA1a基因番茄通过上调Hsp17等基因的表达显著提高抗热性 [6] ,通过上调ATGs基因(autophagy-related,自噬相关基因)的表达显著提高其抗旱性 [30] 。拟南芥中超表达百合HsfA1基因能上调Hsp17.6、18.2、22、70、90.1、101等一系列热激蛋白基因,热激转录因子HsfA2、A7a、A7b基因,转录因子Dreb及甜菜碱合成酶基因Galsyn的表达量 [31] 。拟南芥HsfA2是调控超氧化物清除酶系的关键转录因子,能激活胞质抗坏血酸过氧化物酶(ascorbate peroxidase, APX) APX1和APX2的表达 [18] [32] 。也有少数Hsfs基因被证明是抑制热激蛋白的表达,HsfA5是HsfsA4激活因子的特异抑制因子,它不直接调控热激蛋白的表达,而是通过形成HsfA4/A5复合体使HsfA4不能发挥作用,从而间接抑制热激蛋白基因的表达 [33] 。HsfA1d和HsfA1e是HsfA2的抑制因子,超表达HsfA1d或者HsfA1e都会抑制HsfA2下游基因的表达 [33] 。Xue等从小麦基因组中鉴定了56条Hsfs基因,其中HsfA2b和HsfA4e能转录激活热激蛋白Hsp17和Hsp90基因 [12] ,HsfA6f能转录激活一系列强烈响应热胁迫和干旱胁迫的基因表达,如热激蛋白基因Hsp16.8,Hsp17,Hsp17.3,Hsp90,高尔基体抗凋亡蛋白基因TaGAAP (Golgi anti-apoptotic protein),分子伴侣TaRof和二磷酸核酮糖羧化酶活化酶基因TaRCAL (Rubisco activase large isoform)等 [29] 。
5. 热激转录因子对下游基因的调控机理
正常条件的下,Hsfs转录因子以无活性的单体状态存在于细胞质中,当植物遭受高温或其他逆境胁迫时,Hsfs通过疏水的HR-A和HR-B区相互结合形成活性Hsfs三体,活性三体通过保守的DBD结构域准确的识别并结合于下游基因启动子的HSEs元件,激活热激蛋白Hsp (Heat shock proteins)基因、分子伴侣基因或其他下游基因的转录表达 [25] 。Hsp及辅助分子伴侣能够帮助失活的蛋白质重新组装成有活性的高级结构,维护正常的细胞功能 [3] [25] (图2)。
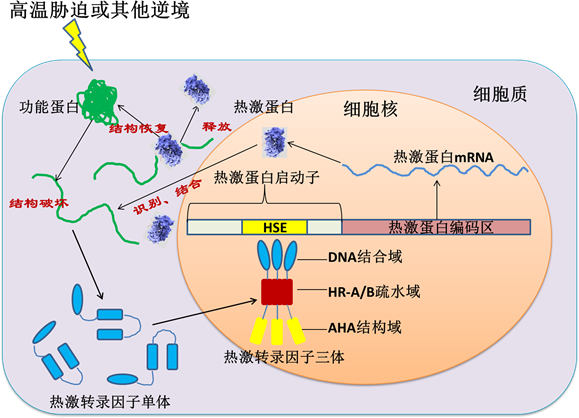
Figure 2. The mechanism of heat shock factors involved in high temperature and other abiotic stress tolerance
图2. 热激转录因子调控植物抵御高温胁迫及其他逆境胁迫的机理
Hsfs通过结合下游基因的启动子中的热激元件HSEs而调控目标基因的表达 [34] 。Hsfs下游基因的启动子中的HSEs具有一致的序列,最初被定义为至少三个重复单位的5’-nGAAn-3’ [34] 。在高等真核生物中,启动子中具有5’-AGAAnnTTCT-3'序列,下游基因才能被Hsfs转录激活 [5] 。在小麦中,通过DNA结合实验,发现TaHsfA2b、TaHsfA4e、TaHsfA6f能接合5’-GAA/Cnn(CTC)/(TTC)/(TTT)nnGAA-3'序列 [12] [29] ,转录激活实验(Transactivation Assay)结果表明,TaHsfA2b、TaHsfA4e、TaHsfA6f对下游基因的激活依赖于HSEs的存在,如果下游基因启动子中删除或者将5’-GAA/Cnn(CTC)/(TTC)/(TTT)nnGAA-3’突变成其他序列,TaHsfA2b、TaHsfA4e、TaHsfA6f则不能转录激活下游基因 [12] [29] 。
6. 展望
随着高温天气和热胁迫灾害的频现,热激转录因子的研究日益成为研究热点。更多的有重要功能的热激转录因子家族成员被挖掘。另外,研究内容日益拓宽,从最开始对胁迫的响应到抗逆性状研究,以及抗逆性机制的研究,研究深度不断加大。目前研究不仅关注与热激因子的抗热性研究,也关注热激转录因子的多调控网络研究,特别是热激因子的抗旱抗热联合调控研究。随着研究的深入,植物抗高温、干热风等非生物逆境胁迫伤害的机理将进一步被揭示。
基金项目
国家大学生创新创业项目(No. 201610452030);国家自然基金青年项目(No. 31000744);山东省自然基金(No. BS2010SW004; No. ZR2016CQ26);山东省重点研发公益类项目(No. 2017NC210010)。
文章引用
赵明宣,胡晓君. 植物热激转录因子研究进展
The Research Progress of Heat Shock Transcription Factors in Plants[J]. 植物学研究, 2018, 07(02): 158-163. https://doi.org/10.12677/BR.2018.72021
参考文献
- 1. Liu, B., Liu, L.L., Tian, L.Y., Cao, W.X., Zhu, Y. and Asseng, S. (2014) Post-Heading Heat Stress and Yield Impact in Winter Wheat of China. Global Change Biology, 20, 372-381.
https://doi.org/10.1111/gcb.12442 - 2. 杨晓娟, 居辉, 王治世, 郭安廷, 杨佑明. 花后高温和干旱对冬小麦光合、抗氧化特性及粒重的影响[J]. 麦类作物学报, 2015, 35(7): 958-963.
- 3. AI-Whaibi, M.H. (2011) Plant Heat-Shock Proteins: A Mini Review. Journal of King Saud University—Science, 23, 139-150.
- 4. Ahuja, I, de Vos, R.C.H., Bones, A.M. and Hall, R.D. (2010) Plant Molecular Stress Responses Face Climate Change. Trends in Plant Science, 15, 664-674.
https://doi.org/10.1016/j.tplants.2010.08.002 - 5. Nover, L., Bharti, K., Döring, P., Mishra, S.K., Ganguli, A. and Scharf, K.-D. (2001) Arabidopsis and the Heat Stress Transcription Factor World: How Many Heat Stress Transcription Factors Do We Need? Cell Stress and Chaperones, 6, 177-189.
https://doi.org/10.1379/1466-1268(2001)006<0177:AATHST>2.0.CO;2 - 6. Mishra, S.K., Tripp, J. and Win-kelhaus, S. (2002) In the Complex Family of Heat Stress Transcription Factors, HsfA1 Has a Unique Role as Master Regulator of Thermotolerance in Tomato. Genes and Development, 16, 1555-1567.
https://doi.org/10.1101/gad.228802 - 7. Reddy, P.S., KaviKishor, P.B., Seiler, C., Kuhlmann, M., Eschen-Lippold, L. and Lee, J. (2014) Unravelling Regulation of the Small Heat Shock Proteins by the Heat Shock Factor HvHsfB2c in Barley: Its Implications in Drought Stress Response and Seed Development. PlosOne, 9, 1371-1397.
https://doi.org/10.1371/journal.pone.0089125 - 8. Guo, J., Wu, J., Ji, Q., Wang, C., Luo, L., Yuan, Y., Wang, Y. and Wang, J. (2008) Genome-Wide Analysis of Heat Shock Transcription Factor Families in Rice and Arabidopsis. Journal of Genetics and Genomics, 35, 105-118.
https://doi.org/10.1016/S1673-8527(08)60016-8 - 9. Jin, G.H., Gho, H.J. and Jung, K.H. (2013) A Systematic View of Rice Heat Shock Transcription Factor Family Using Phylogenomic Analysis. Journal of Plant Physiology, 170, 321-330.
https://doi.org/10.1016/j.jplph.2012.09.008 - 10. Li, P.S., Yu, T.F., He, G.H., Chen, M., Zhou, Y.B., Chai, S.C., Xu, Z.S. and Ma, Y.Z. (2014) Genome-Wide Analysis of the Hsf Family in Soybean and Functional Identification of GmHsf-34 Involvement in Drought and Heat Stresses. BMC Genomics, 21, 1009-1025.
https://doi.org/10.1186/1471-2164-15-1009 - 11. Wang, J., Sun, N., Deng, T., Zhang, L.D. and Zuo, K.K.J. (2014) Genome-Wide Cloning, Identification, Classification and Functional Analysis of Cotton Heat Shock Transcription Factors in Cotton (Gossypium hirsutum). BMC Genomics, 15, 961-980.
https://doi.org/10.1186/1471-2164-15-961 - 12. Xue, G.P., Sadat, S., Drenth, J. and McIntyre, C.L. (2014) The Heat Shock Factor Family from Triticumaestivumin Response to Heat and Other Major Abiotic Stresses and Their Role in Regulation of Heat Shock Protein Genes. Journal of Experimental Botany, 65, 539-557.
https://doi.org/10.1093/jxb/ert399 - 13. Driedonks, N., Xu, J., Peters, J.L., Park, S. and Rieu, I. (2015) Multi-Level Interactions between Heat Shock Factors, Heat Shock Proteins, and the Redox System Regulate Acclimation to Heat. Frontiers in Plant Science, 6, 999-1012.
https://doi.org/10.3389/fpls.2015.00999 - 14. Lohmann, C., Eggers-Schumacher, G., Wunderlich, M. and Schöffl, F. (2004) Two Different Heat Shock Factors Regulate Immediate Early Expression of Stress Genes in Arabidopsis. Molecular Genetics and Genomics, 271, 11-21.
https://doi.org/10.1007/s00438-003-0954-8 - 15. Zhu, B., Ye, C., Lü, H., Chen, X., Chai, G., Chen, J. and Wang, C. (2006) Identification and Characterization of a Novel Heat Shock Transcription Factor Gene, GmHsfA1, in Soybeans (Glycine max). Journal of Plant Research, 119, 247-256.
https://doi.org/10.1007/s10265-006-0267-1 - 16. Nishizawa, A., Yabuta, Y. and Shigeoka, S. (2008) Galactinol and Raffinose Constitute a Novel Function to Protect Plants from Oxidative Damage. Plant Physiology, 147, 1251-1263.
https://doi.org/10.1104/pp.108.122465 - 17. Nishizawa, A., Yabuta, Y., Yoshida, E., Maruta, T., Yoshimura, K. and Shigeoka, S. (2006) Arabidopsis Heat Shock Transcription Factor A2 as a Key Regulator in Response to Several Types of Environmental Stress. The Plant Journal, 48, 535-547.
https://doi.org/10.1111/j.1365-313X.2006.02889.x - 18. Banti, V., Mafessoni, F., Loreti, E., Alpi, A. and Perata, P. (2010) The Heat-Inducible Transcription Factor HsfA2 Enhances Anoxia Tolerance in Arabidopsis. Plant Physiology, 152, 1471-1483.
https://doi.org/10.1104/pp.109.149815 - 19. Yokotani, N., Ichikawa, T., Kondou, Y., Matsui, M., Hirochika, H., Iwabuchi, M. and Oda, K. (2008) Expression of Rice Heat Stress Transcription Factor OsHsfA2e Enhances Tolerance to Environmental Stresses in Transgenic Arabidopsis. Planta, 227, 957-967.
https://doi.org/10.1007/s00425-007-0670-4 - 20. Li, Z., Zhang, L., Wang, A., Xu, X. and Li, J. (2013) Ectopic Overexpression of SlHsfA3, a Heat Stress Transcription Factor from Tomato, Confers Increased Thermotolerance and Salt Hypersensitivity in Germination in Transgenic Arabidopsis. PLoS ONE, 8, 1371-1387.
https://doi.org/10.1371/journal.pone.0054880 - 21. Schmidt, R., Schippers, J.H.M., Welker, A., Mieulet, D., Guiderdoni, E. and Mueller-Roeber, B. (2009) Transcription Factor OsHsfC1b Regulates Salt Tolerance and Develop-ment in Oryza sativa ssp. Japonica. Aob Plants, 11, 1-17.
- 22. Busch, W., Wunderlich, M. and Schöffl, F. (2005) Identification of Novel Heat Shock Factor-Dependent Genes and Biochemical Pathways in Arabidopsis thaliana. The Plant Journal, 41, 1-14.
https://doi.org/10.1111/j.1365-313X.2004.02272.x - 23. Schramm, F., Larkindale, J., Kiehlmann, E., Ganguli, A., Englich, G., Vierling, E. and Von Koskull-Döring, P. (2008) A Cascade of Transcription Factor DREB2A and Heat Stress Transcription Factor HsfA3 Regulates the Heat Stress Response of Arabidopsis. The Plant Journal, 53, 264-274.
https://doi.org/10.1111/j.1365-313X.2007.03334.x - 24. Kumar, M., Busch, W., Birke, H., Kemmerling, B., Nürnberger, T. and Schöffl, F. (2009) Heat Shock Factors HsfB1 and HsfB2b Are Involved in the Regulation of Pdf1.2 Expression and Pathogen Resistance in Arabidopsis. Molecular Plant, 2, 152-165.
https://doi.org/10.1093/mp/ssn095 - 25. Scharf, K.D., Berberich, T., Ebersberger, I. and Nover, L. (2012) The Plant Heat Stress Transcription Factor (Hsf) Family: Structure, Function and Evolution. Biochimicaet Biophysica Acta, 1819, 104-119.
https://doi.org/10.1016/j.bbagrm.2011.10.002 - 26. Baniwal, S.K., Chan, K.Y., Scharf, K.D. and Nover, L. (2007) Role of Heat Stress Transcription Factor HsfA5 as Specific Repressor of HsfA4. The Journal of Biological Chemistry, 282, 3605-3613.
https://doi.org/10.1074/jbc.M609545200 - 27. Ikeda, M., Mitsuda, N. and Ohme-Takagi, M. (2011) Arabidopsis HsfB1 and HsfB2b Act as Repressors of the Expression of Heat-Inducible Hsfs But Positively Regulate the Acquired Thermotolerance. Plant Physiology, 157, 1243-1254.
https://doi.org/10.1104/pp.111.179036 - 28. Charng, Y.Y., Liu, H.C., Liu, N.Y., Chi, W.T., Wang, C.N. and Chang, S.H. (2007) A Heat-Inducible Transcription Factor, HsfA2, Is Required for Extension of Acquired Thermotolerance in Arabidopsis. Plant Physiology, 143, 251-262.
https://doi.org/10.1104/pp.106.091322 - 29. Xue, G.P., Drenth, J. and McIntyre, C.L. (2015) TaHsfA6f Is a Transcriptional Activator That Regulates a Suite of Heat Stress Protection Genes in Wheat (Triticum aestivum L.) Including Previously Unknown Hsf Targets. Journal of Experimental Botany, 66, 1025-1039.
https://doi.org/10.1093/jxb/eru462 - 30. Wang, Y., Cai, S., Yin, L., Shi, K., Xia, X., Zhou, Y., Yu, J. and Zhou, J. (2015) Tomato HsfA1a Plays a Critical Role in Plant Drought Tolerance by Activating ATG Genes and Inducing Autophagy. Autophagy, 11, 2033-2047.
https://doi.org/10.1080/15548627.2015.1098798 - 31. Gong, B., Yi, J., Wu, J., Sui, J., Khan, M.A., Wu, Z., Zhong, X., Seng, S., He, J. and Yi, M. (2014) LiHsfA1, a Novel Heat Stress Transcription Factor in Lily (Lilium longiflorum), Can Interact with LiHsfA2 and Enhance the Thermotolerance of Transgenic Arabidopsis thaliana. Plant Cell Reporter, 33, 1519-1533.
https://doi.org/10.1007/s00299-014-1635-2 - 32. Li, C., Chen, Q., Gao, X., Qi, B., Chen, N. and Xu, S. (2005) AtHsfA2 Modulates Expression of Stress Responsive Genes and Enhances Tolerance to Heat and Oxidative Stress in Arabidopsis. Science in China, Series C Life Science, 48, 540-550.
https://doi.org/10.1360/062005-119 - 33. Nishizawa-Yokoi, A., Nosaka, R., Hayashi, H., Tainaka, H., Maruta, T., Tamoi, M., Ikeda, M., Ohme-Takagi, M., Yoshimura, K., Yabuta, Y. and Shigeoka, S. (2011) HsfA1d and HsfA1e Involved in the Transcriptional Regulation of HsfA2 Function as Key Regulators for the Hsf Signaling Network in Response to Environmental Stress. Plant Cell Physiology, 52, 933-945.
https://doi.org/10.1093/pcp/pcr045 - 34. Morimoto, R.I. (1998) Regulation of the Heat Shock Transcriptional Response: Cross Talk between a Family of Heat Shock Factors, Molecular Chaperones, and Negative Regulators. Genes and Development, 12, 3788-3796.
https://doi.org/10.1101/gad.12.24.3788