Advances in Clinical Medicine
Vol.
10
No.
11
(
2020
), Article ID:
38807
,
10
pages
10.12677/ACM.2020.1011410
基于微阵列数据的胃癌相关核心基因的挖掘和鉴定分析
郭鹏1,赵建刚2,刘义粉2,赵光远2,鲍双振2,刘防震2,尹长恒2,裴晓露3*
1河北医科大学第四医院骨科,河北 石家庄
2哈励逊国际和平医院普外一科,河北 衡水
3河北省人民医院肿瘤内科,河北 石家庄
收稿日期:2020年11月2日;录用日期:2020年11月19日;发布日期:2020年11月26日

摘要
背景:胃癌(GC)是起源于胃黏膜上皮的恶性肿瘤,在我国恶性肿瘤死亡率中占第3位,早期症状不明显,发现时多已发展至晚期,预后较差。为了筛选GC发生发展的潜在基因,本研究从GEO数据库中获得GSE109476和GSE118916进行生物信息学分析。方法:首先,利用GEO2R识别差异表达基因(DEGs),通过GO和KEGG分析对DEGs进行功能注释。利用STRING工具构建蛋白–蛋白相互作用网络,挖掘出最重要的模块和核心基因。结果:共鉴定出229个DEGs。DEGs的功能变化主要集中在细胞黏附、细胞骨架、细胞外基质和胶原蛋白合成等方面。COL4A1、DCN、FSTL1、SPARC、SERPINH1基因被鉴定为核心基因。结论:综上所述,本研究中发现的DEGs和核心基因有可能成为潜在的诊断和治疗靶点。
关键词
胃癌,差异表达基因,蛋白质互作网络,生物信息学

Mining and Identification of Gastric Cancer Related Core Genes Based on Microarray Data
Peng Guo1, Jiangang Zhao2, Yifen Liu2, Guangyuan Zhao2, Shuangzhen Bao2, Fangzhen Liu2, Changheng Yin2, Xiaolu Pei3*
1Department of Orthopedics, The Fourth Hospital of Hebei Medical University, Shijiazhuang Hebei
2Department of General Surgery I, Harrison International Peace Hospital, Hengshui Hebei
3Department of Medical Oncology, Hebei General Hospital, Shijiazhuang Hebei
Received: Nov. 2nd, 2020; accepted: Nov. 19th, 2020; published: Nov. 26th, 2020

ABSTRACT
Background: Gastric cancer (GC) is a malignant tumor originating from gastric mucosal epithelium, accounting for the third place in the mortality rate of malignant tumors in China. The early symptoms are not obvious, and most of them have advanced stage at the time of discovery, with poor prognosis. In order to screen the potential genes for GC occurrence and development, this study obtained GSE109476 and GSE118916 from the GEO database for bioinformatics analysis. Methods: First, the differentially expressed genes (DEGs) were identified by GEO2R and functional annotation of DEGs was performed by GO and KEGG analysis. The STRING tool was used to construct the protein-protein interaction network, and the most important modules and core genes were mined. Results: A total of 229 DEGs were identified. The functional changes of DEGs mainly focus on the aspects of cell adhesion, cytoskeleton, extracellular matrix and collagen synthesis. COL4A1, DCN, FSTL1, SPARC and SERPINH1 genes were identified as core genes. Conclusion: In summary, the DEGs and core genes identified in this study may be potential diagnostic and therapeutic targets.
Keywords:Gastric Cancer, Differentially Expressed Genes, Protein Interaction Network, Bioinformatics

Copyright © 2020 by author(s) and Hans Publishers Inc.
This work is licensed under the Creative Commons Attribution International License (CC BY 4.0).
http://creativecommons.org/licenses/by/4.0/


1. 引言
胃癌系指源于胃黏膜上皮细胞的恶性肿瘤,主要是胃腺癌。目前,胃癌在全球发病率位居第四位、病死率位居第二位,是严重危害人类健康的恶性肿瘤之一 [1]。胃癌的发生与人群不良环境、生活方式、饮食结构的改变以及幽门螺旋菌感染具有密切的联系,早期胃癌症状不明显,部分患者可有消化不良症状,晚期胃癌可有上腹痛、餐后加重、纳差、厌食、乏力及体重减轻等症状。待患者出现明显症状时入院检查,疾病多已发展至胃癌晚期,丧失最佳手术治疗时机。如果能早期诊断出胃癌,其5年生存率将上升到95% [2],这提示着改善胃癌预后的根本方法在于早发现、早诊断、早治疗。目前一些血清生物标志物被用于早期胃癌的筛查,如CA19-9和CEA,但是这些肿瘤标志物的敏感性和特异性较低 [3]。因此,寻找一种新的有效的早期胃癌生物标志物,进一步探索胃癌的发病机制,以寻找潜在的诊断和治疗靶点,实现早发现、早诊断与靶向治疗,具有重要的临床意义。
生物信息学是将生命科学和计算机科学结合的新兴的交叉学科,主要研究生物信息的采集、存储、处理、传播、分析和解释。通过利用生物和信息学技术能够处理分析大量复杂的生物数据。微阵列数据信息分析技术已经广泛应用于肿瘤等疾病的研究,以探索其中的遗传相关性 [4] [5]。微阵列分析技术能够同时获取数万个基因的表达信息,进而发掘与疾病发生发展相关的基因组学改变。目前已有大量研究 [6] [7] 使用生物信息学技术来分析肿瘤进展中的差异表达基因,进而研究其在生物过程、分子功能和信号通路方面的作用,并阐明疾病的发病机制,为早期诊断及治疗提供理论依据。
本研究应用生物信息学技术,对比胃癌患者标本与正常人的基因数据,进而筛选差异表达基因和核心基因。这些发现将提高我们对胃癌的潜在机制的认识,为寻找新的诊断标志物及靶向治疗提供基础。
2. 材料和方法
2.1. 数据集
我们从gene Expression Omnibus (GEO) (https://www.ncbi.nlm.nih.gov/geo/)数据库下载了GSE109476和GSE118916两个基因表达谱。GSE109476包括5个胃癌样本和5个正常样本,GSE118916包括15个胃癌样本和15个正常样本。
2.2. DEGs的鉴别
我们使用了GEO2R (http://www.ncbi.nlm.nih.gov/geo/geo2r),这是一个基于GEOquery和limma R包的在线工具来识别胃癌和正常组之间的差异表达基因。截止条件是P-value < 0.05和log (FC) >1或log (FC) < −1。
2.3. DEGs注释
DAVID (https://david.ncifcrf.gov/home.jsp)和Metascape (http://metascape.org/gp/index.html)是两个功能强大的注释工具,可以对基因进行生物过程(BP)、细胞成分(CC)、分子功能(MF)和KEGG分析。我们通过DAVID和Metascape注释了DEGs的功能。
2.4. 蛋白质互作网络的构建
互作基因检索工具(STRING) (http://string-db.org)可以将DEGs转化为表达蛋白并构建PPI网络。我们通过STRING得到了DEGs的PPI网络,并通过Cytoscape(3.8.0版本)将其可视化。
2.5. 关键基因的鉴别和表达
使用Cytoscape的开放插件——MCODE(1.6.1版)来识别PPI网络中的重要模块。评分标准:the maximum depth = 100,MCODE scores > 5,cut-off = 2,k-score = 2,node score cut-off = 0.2。此外,我们还利用cytoHubb从MCODE的关键模块中筛选出核心基因,并通过MCC和DMNC对其进行排序。
3. 结果
3.1. 差异表达基因
两个火山图呈现出GSE109476和GSE118916中的DEGs (图1(A)、图1(B))。维恩图显示了两个数据集共有229个DEGs (图2)。
3.2. 差异表达基因的注释
Metascape的富集分析结果如图3所示。分别以气泡图显示DEGs相关的生物过程(BP)、细胞成分(CC)、分子功能(MF)和KEGG富集分析的结果(图4(A)~(D))。
3.3. 蛋白质互作网络和核心基因
PPI结果如图5(A)所示。MCODE分析的关键模块如图5(B)所示。通过cytoHubb分析筛选出的前10个基因如图5(C),图5(D)所示。Veen diagram计算出了算法之间的四个相互基因(图6)。两张热图分别显示了核心基因在GSE109476 (图7(A))和GSE118916 (图7(B))中的表达情况。
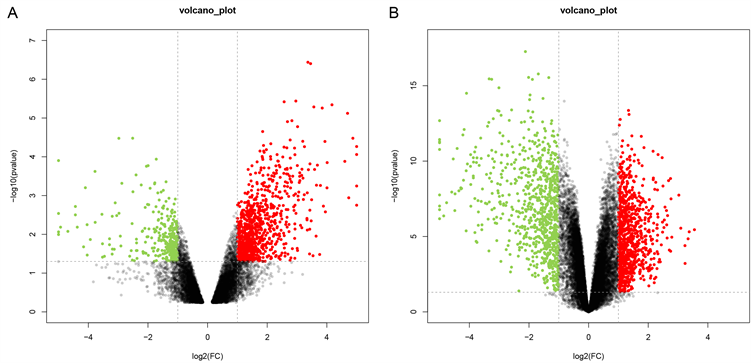
Figure 1. Two volcano plots present the DEGs. (A) The DEGs in the GSE109476. (B) The DEGs in the GSE118916
图1. 两个火山图呈现差异表达基因。(A) GSE109476中的差异表达基因;(B) GSE118916中的差异表达基因
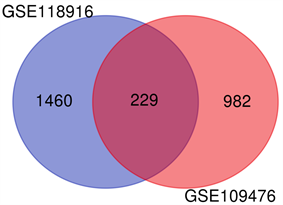
Figure 2. The common DEGs between GSE109476 and GSE118916
图2. GSE109476和GSE118916中共有的差异表达基因
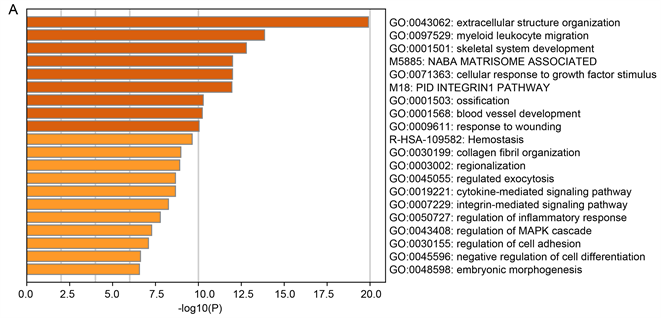
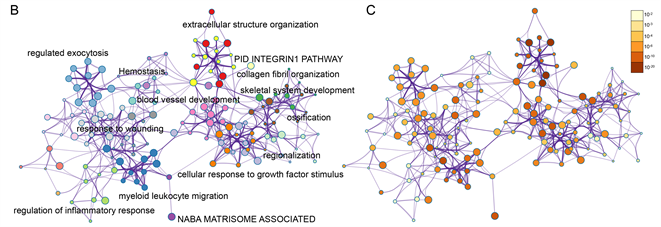
Figure 3. Enrichment analysis for the DEGs by Metascape. (A) Heatmap of enriched terms across input differently expressed gene lists, colored by p-values, via the Metascape. (B) Network of enriched terms colored by cluster identity, where nodes that share the same cluster identity are typically close to each other. (C) Network of enriched terms colored by p-value, where terms containing more genes tend to have a more significant p-value
图3. 通过Metascape做差异表达基因的富集分析。(A) 在Metascape中,通过输入差异表达表达基因列表的丰富项热图,用p值着色;(B) 用集群标识着色的富集术语的网络,其中共享相同集群标识的节点通常彼此靠近。(C) 由p值着色的富集术语网络,其中包含更多基因的术语的p值往往更显著
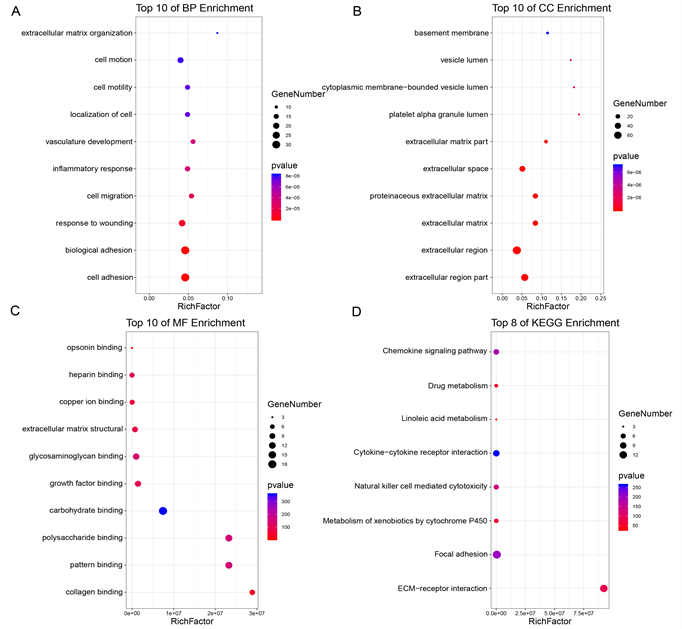
Figure 4. The functional annotation for the DEGs based on the DAVID. (A) BP, (B) CC, (C) MF, and (D) KEGG
图4. 基于DAVID,差异表达基因的功能注释。(A) 生物过程;(B) 细胞组分;(C) 分子功能;(D) 通路
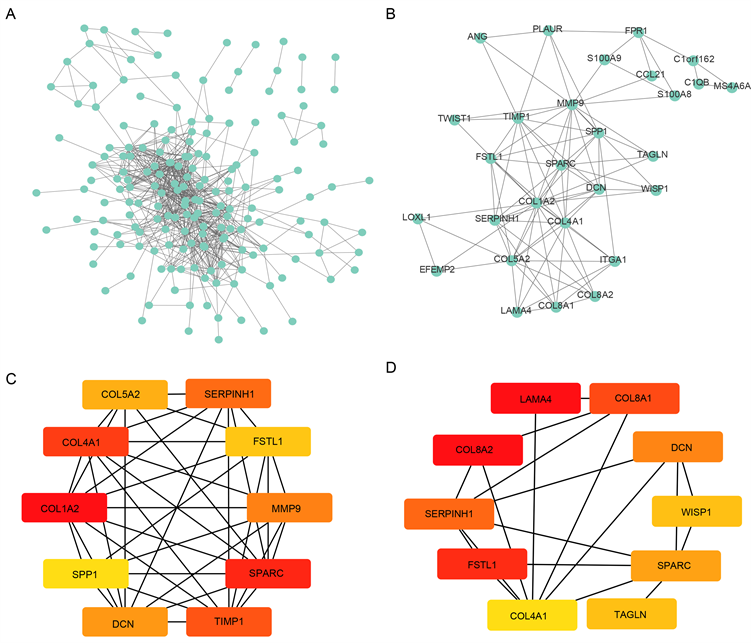
Figure 5. The PPI network and hub genes. (A) PPI network; (B) The key module of MCODE analysis; (C) The top 10 genes screened by cytoHubb in MCC; (D) The top 10 genes screened by cytoHubb in DMNC
图5. 蛋白–蛋白互作网络分析和核心基因。(A) 蛋白–蛋白互作网络分析;(B) MCODE分析;(C) 通过cytoHubb中MCC算法找到的10个核心基因;(D) 通过cytoHubb中DMNC算法找到的10个核心基因
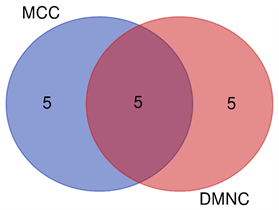
Figure 6. Venn diagram figured out five mutual genes between the algorithms, which included SERPINH1, SPARC, FSTL1, DCN, COL4A1
图6. 韦恩图显示两种算法之间的5个共有基因,包含:SERPINH1, SPARC, FSTL1, DCN, COL4A1
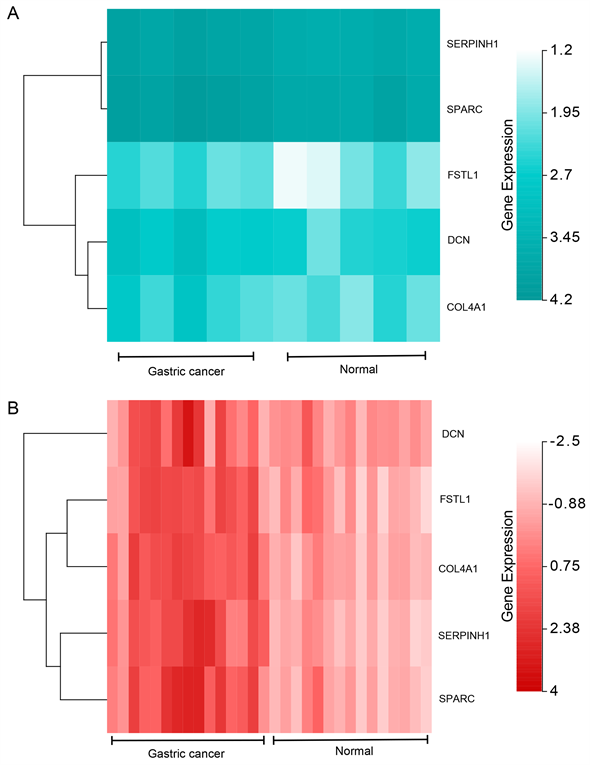
Figure 7. The expression analysis for the hub genes. (A) The heat map showed the expressions of the hub genes in GSE109476; (B) The heat map showed the expressions of the hub genes in GSE118916
图7. 核心基因的表达量分析。(A) 热图显示出GSE109476中核心基因的表达情况;(B) 另一个热图显示出GSE118916中核心基因的表达情况
4. 讨论
胃癌是一种具有高度异质性的恶性肿瘤,其发病机制主要与遗传物质改变、幽门螺杆菌及EB病毒(Epstein-Barr virus, EBV)感染及表观遗传学改变相关 [8] [9] [10]。目前,手术切除后辅以化疗是胃癌患者常规治疗手段之一,一系列化疗药物和分子靶向药物,如奥沙利铂、5-氟尿嘧啶及赫赛汀等 [11] 提高胃癌患者的临床治疗效果,但由于其普遍存在药物毒副作用,胃癌患者的预后水平仍不容乐观 [12]。因此,有必要寻找新的胃癌分子治疗靶点并探讨其与胃癌预后的相关性,进一步优化胃癌患者的精准治疗。
生物信息学技术已经被广泛用于寻找与肿瘤发生发展相关的遗传分子,进而找到可以作为治疗靶点的基因和分子。Cao等通过该技术找到与胃癌相关的分子PLEKHG1,通过进一步验证证实该基因与胃癌的相关性,提示该分子作为诊断和预测转归的生物标志物 [13]。Wang等通过生信技术找到与结直肠癌增殖和转移相关的分子Linc01194,提示其可能作为潜在治疗靶点 [14]。本研究通过分析2个mRNA微阵列数据集,获得胃癌组织与非癌组织间的DEGs。在2个数据集中共鉴定出229个DEGs。通过生信分析发现胃癌患者中COL4A1、DCN、FSTL1、SPARC、SERPINH1高表达。同时,通过Gene Ontology (GO)分析和Kyoto Encyclopedia of Genes and Genomes (KEGG)分析及Gene Set Enrichment Analysis (GSEA)找到多个明显上调及下调的gene set。
COL4A1 (collagen type IV alpha 1,即Ⅳ型胶原蛋白α1)属于IV型胶原蛋白家族,包含三个结构域,是构成基底膜的重要成分 [15]。近期研究发现COL4A1与肿瘤侵袭、转移等生物学进程密切相关。Jin等 [16] 研究发现,与正常乳腺上皮细胞相比,乳腺浸润性导管癌中存在478个差异表达基因;功能富集分析结果表明,大部分差异表达基因与ECM受体相互作用有关;进一步实验结果证实,下调COL4A1基因的表达可以显著抑制乳腺浸润性导管癌细胞的集落形成,抑制癌细胞的增殖能力。COL4A1基因作为关键枢纽基因,在浸润性导管癌的增殖和集落形成中起重要作用。Miyake等 [17] 运用原位肿瘤模型分析人尿路上皮癌细胞系MGH-U3、UM-UC-14和UM-UC-3显示的结节、小梁和浸润模式,并通过基因微阵列芯片鉴定出COL4A1和COL13A1两种胶原蛋白,siRNA靶向干扰COL4A1和COL13A1可显著抑制肿瘤浸润,肿瘤细胞产生的COL4A1和COL13A1通过诱导肿瘤出芽,在肿瘤侵袭中起关键作用。Miyake等 [18] 研究发现,尿液中COL4A1和COL13A1高表达是尿路上皮癌患者膀胱内复发的独立危险因素。Huang等 [19] 采用生物信息学方法预测到COL4A1在曲妥珠单抗耐药胃癌患者中的mRNA表达水平较未耐药者显著升高,提示COL4A1高表达可诱导胃癌患者耐药。本研究发现COL4A1在胃癌组织中高表达,且较低表达者预后差。同样有两项生物信息学挖掘研究 [20] [21] 均揭示COL4A1在胃癌中高表达,并与患者预后密切相关,与本研究的结果一致。COL4A1高表达可以促进胃癌的增殖、侵袭和迁移能力,而低表达的细胞可抑制胃癌细胞增殖,延缓细胞迁移,因此COL4A1可以作为一种潜在的生物标志物和治疗靶点。
FSTL1 (卵泡抑素样蛋白1)是由多种细胞分泌的分泌性细胞外糖蛋白,主要由间充质来源的细胞产生,其在人体组织中广泛表达,在细胞生存、增殖、分化、迁移和胚胎脏器成熟过程中起着重要的调节作用 [22]。其编码与卵泡抑素相似的蛋白,可激活NF-κB并参与TGF-beta/BMP信号通路,调节细胞的分化与凋亡 [23]。Bae等研究发现敲除FSTL1基因可诱导肺癌细胞凋亡 [24]。相反,Chan等研究发现FSTL1可以抑制卵巢癌和子宫内膜癌的发展 [25]。张等 [26] 的体外实验结果表明,FSTL1可促进肺癌、乳腺癌等多种恶性肿瘤细胞生长。FSTL1在不同类型的肿瘤中发挥的作用完全相反,可能与他们都参与在肿瘤发生发展的不同阶段发挥截然相反作用的TGF-beta相关信号通路有关 [27]。目前FSTL1与胃癌的关系的研究尚少,本研究发现其在胃癌组织中高表达,且预后较差,它可能有促进胃癌发生发展的作用。
SPARC (Secreted Protein Acidic And Cysteine Rich,富含半胱氨酸的酸性分泌蛋白)是一种相对分子质量32,000的细胞外非结构分泌基质糖蛋白,由一个单一的多肽(285个氨基酸)组成,最早由美国TERMINE等于1981年在人和胎牛骨中分离提纯 [28]。研究发现,在一些具有高转移特征的肿瘤中,如胶质母细胞瘤、黑色素瘤、乳腺癌和前列腺癌,SPARC能促进骨转移和上皮–间质转化(EMT),促进肿瘤的发展,但在胰腺癌、结直肠癌、胃癌等低转移性肿瘤中可作为抗血管生成、促凋亡、抑制细胞增殖和抑制细胞周期的抗肿瘤因子 [29] [30]。其在GC细胞中的作用备受争议。Tsutomu等 [31] 研究发现与正常对照组相比,胃癌组织中SPARC mRNA表达水平升高,且与低SPARC表达相比,高SPARC表达预后较差。Chen等 [32] 也表明,在140名卵巢癌患者中,高SPARC表达与低SPARC表达相比,其预后更差。而Chew等和Liang等分别报道了SPARC低表达与120例和114例结直肠癌患者较差的长期生存相关 [33] [34]。SPARC可能在不同的癌症和同一癌症的不同发展阶段发挥不同的作用。本研究发现在胃癌组织中SPARC表达高于癌旁组织,且预后较差。
尽管本研究进行了严谨地生物信息学分析,但其中仍有不足:1) 数据集中样本量较少,需进一步扩大样本量来得到更为准确的结果。2) 需用大量的临床样本、动物实验进行综合验证,以便更加深入地了解原发性结直肠癌的发病机制。
总之,我们通过生物信息分析,从胃癌患者与正常组织基因样本中鉴定出了5个明显差异表达基因。DEGs中的关键基因可能为胃癌的诊断及靶向治疗提供新的思路及证据。
文章引用
郭 鹏,赵建刚,刘义粉,赵光远,鲍双振,刘防震,尹长恒,裴晓露. 基于微阵列数据的胃癌相关核心基因的挖掘和鉴定分析
Mining and Identification of Gastric Cancer Related Core Genes Based on Microarray Data[J]. 临床医学进展, 2020, 10(11): 2695-2704. https://doi.org/10.12677/ACM.2020.1011410
参考文献
- 1. Digklia, A. and Wagner, A.D. (2016) Advanced Gastric Cancer: Current Treatment Landscape and Future Perspectives. World Journal of Gastroenterology, 22, 2403-2414. https://doi.org/10.3748/wjg.v22.i8.2403
- 2. Parkin, D.M., Bray, F., Ferlay, J. and Pisani, P. (2005) Global Cancer Statistics, 2002. CA: A Cancer Journal for Clinicians, 55, 74-108. https://doi.org/10.3322/canjclin.55.2.74
- 3. Shimada, H., Noie, T., Ohashi, M., Oba, K. and Takahashi, Y. (2014) Clinical Significance of Serum Tumor Markers for Gastric Cancer: A Systematic Review of Literature by the Task Force of the Japanese Gastric Cancer Association. Gastric Cancer, 17, 26-33. https://doi.org/10.1007/s10120-013-0259-5
- 4. Zhou, Y., Liepe, J., Sheng, X., Stumpf, M.P. and Barnes, C. (2011) GPU Accelerated Biochemical Network Simulation. Bioinformatics, 27, 874-876. https://doi.org/10.1093/bioinformatics/btr015
- 5. Nobile, M.S., Cazzaniga, P., Tangherloni, A. and Besozzi, D. (2017) Graphics Processing Units in Bioinformatics, Computational Biology and Systems Biology. Briefings in Bioinformatics, 18, 870-885. https://doi.org/10.1093/bib/bbw058
- 6. Chaudhary, K., Poirion, O.B., Lu, L. and Garmire, L.X. (2018) Deep Learning-Based Multi-Omics Integration Robustly Predicts Survival in Liver Cancer. Clinical Cancer Research, 24, 1248-1259. https://doi.org/10.1158/1078-0432.CCR-17-0853
- 7. Qiu, Z., Li, H., Zhang, Z., et al. (2019) A Pharmacogenomic Landscape in Human Liver Cancers. Cancer Cell, 36, 179-193.e11. https://doi.org/10.1016/j.ccell.2019.07.001
- 8. Hoshi, H. (2020) Management of Gastric Adenocarcinoma for General Surgeons. Surgical Clinics of North America, 100, 523-534. https://doi.org/10.1016/j.suc.2020.02.004
- 9. Wadhwa, R., Song, S., Lee, J.S., Yao, Y., Wei, Q. and Ajani, J.A. (2013) Gastric Cancer-Molecular and Clinical Dimensions. Nature Reviews Clinical Oncology, 10, 643-655. https://doi.org/10.1038/nrclinonc.2013.170
- 10. Song, S. and Ajani, J.A. (2013) The Role of microRNAs in Cancers of the Upper Gastrointestinal Tract. Nature Reviews Gastroenterology & Hepatology, 10, 109-118. https://doi.org/10.1038/nrgastro.2012.210
- 11. Mongan, A.M., Kalachand, R., King, S., et al. (2015) Outcomes in Gastric and Junctional Cancer Using Neoadjuvant and Adjuvant Chemotherapy (Epirubicin, Oxaliplatin, and Capecitabine) and Radical Surgery. Irish Journal of Medical Science, 184, 417-423. https://doi.org/10.1007/s11845-014-1135-y
- 12. Karimi, P., Islami, F., Anandasabapathy, S., Freedman, N.D. and Kamangar, F. (2014) Gastric Cancer: Descriptive Epidemiology, Risk Factors, Screening, and Prevention. Cancer Epidemiology, Biomarkers & Prevention, 23, 700-713. https://doi.org/10.1158/1055-9965.EPI-13-1057
- 13. Cao, W., Zhou, D., Tang, W., An, H. and Zhang, Y. (2019) Discovery of Plasma Messenger RNA as Novel Biomarker for Gastric Cancer Identified through Bioinformatics Analysis and Clinical Validation. PeerJ, 7, e7025. https://doi.org/10.7717/peerj.7025
- 14. Wang, X., Liu, Z., Tong, H., et al. (2019) Linc01194 Acts as an Oncogene in Colorectal Carcinoma and Is Associated with Poor Survival Outcome. Cancer Management and Research, 11, 2349-2362. https://doi.org/10.2147/CMAR.S189189
- 15. Meuwissen, M.E., Halley, D.J., et al. (2015) The Expanding Phenotype of COL4A1 and COL4A2 Mutations: Clinical Data on 13 Newly Identified Families and a Review of the Literature. Genetics in Medicine, 17, 843-853. https://doi.org/10.1038/gim.2014.210
- 16. Jin, R., Shen, J., Zhang, T., et al. (2017) The Highly Expressed COL4A1 Genes Contributes to the Proliferation and Migration of the Invasive Ductal Carcinomas. Oncotarget, 8, 58172-58183. https://doi.org/10.18632/oncotarget.17345
- 17. Miyake, M., Hori, S., Morizawa, Y., et al. (2017) Collagen Type IV Alpha 1 (COL4A1) and Collagen Type XIII Alpha 1 (COL13A1) Produced in Cancer Cells Promote Tumor Budding at the Invasion Front in Human Urothelial Carcinoma of the Bladder. Oncotarget, 8, 36099-36114. https://doi.org/10.18632/oncotarget.16432
- 18. Miyake, M., Morizawa, Y., et al. (2017) Diagnostic and Prognostic Role of Urinary Collagens in Primary Human Bladder Cancer. Cancer Science, 108, 2221-2228. https://doi.org/10.1111/cas.13384
- 19. Huang, R., Gu, W., Sun, B. and Gao, L. (2018) Identification of COL4A1 as a Potential Gene Conferring Trastuzumab Resistance in Gastric Cancer Based on Bioinformatics Analysis. Molecular Medicine Reports, 17, 6387-6396. https://doi.org/10.3892/mmr.2018.8664
- 20. Li, T., Gao, X., Han, L., Yu, J. and Li, H. (2018) Identification of Hub Genes with Prognostic Values in Gastric Cancer by Bioinformatics Analysis. World Journal of Surgical Oncology, 16, 114. https://doi.org/10.1186/s12957-018-1409-3
- 21. Cao, L., Chen, Y., Zhang, M., et al. (2018) Identification of Hub Genes and Potential Molecular Mechanisms in Gastric Cancer by Integrated Bioinformatics Analysis. PeerJ, 6, e5180. https://doi.org/10.7717/peerj.5180
- 22. Lee, S.Y., Kim, D.Y., Kyung, K.M., et al. (2019) High Circulating Follistatin-Like Protein 1 as a Biomarker of a Metabolically Unhealthy State. Endocrine Journal, 66, 241-251. https://doi.org/10.1507/endocrj.EJ18-0352
- 23. Wu, J., Dong, Y., Teng, X., Cheng, M., Shen, Z. and Chen, W. (2015) Follistatin-Like 1 Attenuates Differentiation and Survival of Erythroid Cells through Smad2/3 Signaling. Biochemical and Biophysical Research Communications, 466, 711-716. https://doi.org/10.1016/j.bbrc.2015.09.044
- 24. Bae, K., Park, K.E., Han, J., Kim, J., Kim, K. and Yoon, K.A. (2016) Mitotic Cell Death Caused by Follistatin-Like 1 Inhibition Is Associated with Up-Regulated Bim by Inactivated Erk1/2 in Human Lung Cancer Cells. Oncotarget, 7, 18076-18084. https://doi.org/10.18632/oncotarget.6729
- 25. Chan, Q.K., Ngan, H.Y., Ip, P.P., Liu, V.W., Xue, W.C. and Cheung, A.N. (2009) Tumor Suppressor Effect of Follistatin-Like 1 in Ovarian and Endometrial Carcinogenesis: A Differential Expression and Functional Analysis. Carcinogenesis, 30, 114-121. https://doi.org/10.1093/carcin/bgn215
- 26. Zhang, Y., Xu, X., Yang, Y., et al. (2018) Deficiency of Follistatin-Like Protein 1 Accelerates the Growth of Breast Cancer Cells at Lung Metastatic Sites. Journal of Breast Cancer, 21, 267-276. https://doi.org/10.4048/jbc.2018.21.e43
- 27. Massagué, J. (2008) TGFbeta in Cancer. Cell, 134, 215-230. https://doi.org/10.1016/j.cell.2008.07.001
- 28. Termine, J.D., Kleinman, H.K., Whitson, S.W., Conn, K.M., McGarvey, M.L. and Martin, G.R. (1981) Osteonectin, a Bone-Specific Protein Linking Mineral to Collagen. Cell, 26, 99-105. https://doi.org/10.1016/0092-8674(81)90037-4
- 29. Feng, J. and Tang, L. (2014) SPARC in Tumor Pathophysiology and as a Potential Therapeutic Target. Current Pharmaceutical Design, 20, 6182-6190. https://doi.org/10.2174/1381612820666140619123255
- 30. Nagaraju, G.P., Dontula, R., El-Rayes, B.F. and Lakka, S.S. (2014) Molecular Mechanisms Underlying the Divergent Roles of SPARC in Human Carcinogenesis. Carcinogenesis, 35, 967-973. https://doi.org/10.1093/carcin/bgu072
- 31. Sato, T., Oshima, T., Yamamoto, N., et al. (2013) Clinical Significance of SPARC Gene Expression in Patients with Gastric Cancer. Journal of Surgical Oncology, 108, 364-368. https://doi.org/10.1002/jso.23425
- 32. Chen, J., Wang, M., Xi, B., et al. (2012) SPARC Is a Key Regulator of Proliferation, Apoptosis and Invasion in Human Ovarian Cancer. PLoS ONE, 7, e42413. https://doi.org/10.1371/journal.pone.0042413
- 33. Chew, A., Salama, P., Robbshaw, A., et al. (2011) SPARC, FOXP3, CD8 and CD45 Correlation with Disease Recurrence and Long-Term Disease-Free Survival in Colorectal Cancer. PLoS ONE, 6, e22047. https://doi.org/10.1371/journal.pone.0022047
- 34. Liang, J.F., Wang, H.K., Xiao, H., et al. (2010) Relationship and Prognostic Significance of SPARC and VEGF Protein Expression in Colon Cancer. Journal of Experimental & Clinical Cancer Research, 29, 71. https://doi.org/10.1186/1756-9966-29-71
NOTES
*通讯作者。